
Induction of two cytochrome P450 genes, Cyp6a2 and Cyp6a8,
of Drosophila melanogaster by caffeine
in adult flies and in cell culture
Srividya Bhaskara, Erika Danielle Dean, Vita Lam, Ranjan Ganguly
⁎
Department of Biochemistry, Cellular and Molecular Biology, University of Tennessee, Knoxville, TN 37996, United States
Received 8 December 2005; received in revised form 7 February 2006; accepted 28 February 2006
Available online 5 April 2006
Received by Igor B. Rogozin
Abstract
To examine whether caffeine, the most widely used xenobiotic compound, would induce insect cytochrome P450 or CYP gene expression,
upstream DNA fragments of Cyp6a2 (0.12, 0.26, 0.52 and 0.98-kb) and Cyp6a8 (0.06, 0.1, 0.2, 0.5 and 0.8-kb) genes of Drosophila melanogaster
were individually fused to the firefly luciferase (luc) reporter gene. Promoter activities of these constructs were examined in Drosophila SL-2 cells
using luciferase assays. Activity of 0.2- and 0.8-kb upstream DNA of Cyp6a8 was also measured in transgenic female flies. When these flies were
treated with 2 mM pure caffeine or Vivarin caffeine, both DNA fragments showed a 4
–5-fold induction of promoter activity. Endogenous Cyp6a8 and
Cyp6a2 genes in these flies also showed caffeine-induced expression. In addition, both 0.2- and 0.8-kb DNAs showed differential basal and caffeine-
induced activity in head, ovaries, gut, cuticle plus fat body and malpighian tubules. However, in all tissues 0.8-kb DNA always showed higher basal
and caffeine-induced activities compared to the 0.2-kb DNA, suggesting that the additional DNA present in the 0.8-kb fragment has sequences that
enhance both activities. In SL-2 cells, all reporter constructs of each Cyp6 gene showed significantly higher basal activity than the empty vector.
Sequences that boost basal activity are located in
−265/−129 and −983/−522 DNA of Cyp6a2, and −199/−109 and −491/−199 DNA of Cyp6a8
genes. While the 0.12- and 0.1-kb upstream DNAs of Cyp6a2 and Cyp6a8 genes respectively did not show caffeine-inducibility in SL-2 cells, the
longest upstream DNA of each gene gave the highest level of induction. Caffeine-responsive sequences are not clustered at one place; they appear to
be dispersed in
−983/−126 and −761/−109 regions of Cyp6a2 and Cyp6a8 genes which also contain many binding sites for activator protein 1 (AP1)
and cyclic AMP response element binding protein (CRE-BP). Significance of these binding sites in caffeine-inducibility has been discussed.
© 2006 Elsevier B.V. All rights reserved.
Keywords: Vivarin; Reporter gene assay; Transgenic flies; Cell transfection; Luciferase assay; Promoter assay; AP1 binding sites
1. Introduction
Cytochrome P450 monooxygenases or CYPs comprise a
superfamily of enzymes that are involved in the biosynthesis of
many biologically important compounds and metabolism of a
variety of xenobiotic (foreign) chemicals to which living organ-
isms are exposed to on a daily basis (
). CYPs
have been studied in various organisms from different biological
perspectives. One such perspective is the induction and
regulation of mammalian and human CYP genes, especially in
relation to the metabolism of different toxic chemicals,
carcinogenic compounds and drugs including barbiturate com-
pounds such as phenobarbital and barbital (
). Interestingly, many of these xenobiotic
compounds have been used as tools to identify the cis-regulatory
elements, receptors and other factors that regulate CYP gene
expression in mammals (
Sueyoshi and Negishi, 2001; Hankin-
).
Besides drugs and environmental toxicants, caffeine is
probably another xenobiotic compound to which most people
are exposed everyday. Caffeine is naturally found in berries,
seeds and leaves of many plants including coffee, cocoa and tea.
Gene 377 (2006) 56
–64
www.elsevier.com/locate/gene
Abbreviations: bp, base pair(s); CYP, cytochrome P450 monooxygenase;
Cyp (or CYP), CYP-encoding gene; kb, kilobase(s) or 1000 bp; LUC,
Luciferase; luc, LUC-encoding gene; nt, nucleotide(s).
⁎ Corresponding author. Tel.: +1 865 974 5148; fax: +1 865 974 6306.
E-mail address:
(R. Ganguly).
0378-1119/$ - see front matter © 2006 Elsevier B.V. All rights reserved.
doi:

A variety of commercial food and drinks also contain a high
quantity of caffeine. Because caffeine is consumed heavily and
it is chemically very similar to other purines, extensive studies
have been done on the effect of caffeine on various biological
processes in humans and other mammals such as brain motor
activity, cognitive functions, sleep and hypertension (
et al., 1999; Lorist and Tops, 2003
). However, not much work
has been done on the effect of caffeine on gene expression
except for three studies. One study showed that caffeine
treatment increases CYP1A1 and CYP1A2 mRNA levels in rat
liver and kidney (
). In the other two studies
caffeine was found to increase the levels of c-fos, c-jun, and
junB mRNAs in rat striatum (
), and
sonic hedgehog mRNA in primary murine neuronal and
astroglial cells in culture (
).
In insects, CYPs are known to be involved in conferring
insecticide resistance, and insecticide resistance-associated
overexpression of one or more CYP genes has been observed
in many insect species (see
for review). In
Drosophila also, at least five Cyp genes, including Cyp6a2
and Cyp6a8, show overexpression in DDT resistant strains
(
Maitra et al., 1996; Dombrowski et al., 1998; Daborn et al.,
). However, the mechanism of
overexpression or the regulation of insect CYP genes in general
is not well-understood. Since xenobiotic compounds have been
successfully used to understand mammalian CYP gene regula-
tion, we surmised whether caffeine could also be used to study
insect CYP gene regulation. Therefore, in the present study we
used Drosophila as a model insect and examined the effect of
caffeine on the Cyp6a2 and Cyp6a8 gene expression in adult fly
and also in Drosophila cells in culture. The results showed that
both genes are induced by caffeine, and Cyp6a8 shows
differential constitutive and caffeine-induced expression in
different tissues of the adult flies.
2. Materials and methods
2.1. Fly strains and cell culture
The 91-C strain and the following five reporter transgenic lines
of Drosophila melanogaster created by
in the
ry
506
host strain were used: 0.2-luc30-4 (also referred as 0.2-luc4),
0.8-luc110, 0.8-luc14, 0.8-luc121 and 3.1-luc2. Each of these
transgenic lines has a single reporter transgene with a firefly
luciferase (luc) reporter gene under the control of 0.2- (
−11/
−199), 0.8- (−11/−761) or 3.1-kb (−11/−3100) upstream DNA of
the Cyp6a8 gene. The chromosomal locations of the transgene in
different lines have been described earlier (
). The
0.2-luc4 and 0.8-luc110 strains were made homozygous for their
2nd chromosome-linked transgene by using appropriate balancer
stocks (
) and two genetic crossing schemes. One
scheme gave 0.2-luc4H and 0.8-luc110H stocks. Genotypes of
these flies are +/+; 0.2-luc/0.2-luc; +/+ and +/+; 0.8-luc/0.8-luc; +/
+, respectively. The transgene carrying 2nd chromosomes of these
stocks are from the ry
506
strain that was used for transformation.
The other scheme produced 0.2-luc4H-ry and 0.8-luc110H-ry
stocks. Besides the transgene carrying 2nd chromosomes, the X
and 3rd chromosomes in these stocks are also from the ry
506
strain
(indicated with r). Genotypes of these stocks are X
r
/X
r
; 0.2-luc/
0.2-luc; 3
r
/3
r
and X
r
/X
r
; 0.8-luc/0.8-luc; 3
r
/3
r
, respectively. The
source of the 4th chromosome in any of these stocks is not known.
Other reporter transgenic lines, 0.8-luc14, 0.8-luc121 and 3.1-
luc2, carrying 3rd chromosome-linked reporter transgenes are the
original transgenic lines made in the ry
506
host strain. These strains
have been maintained by crossing ry
+
transgenic males and
females in every generation. Flies were reared at 25 °C on standard
cornmeal-agar-molasses Drosophila medium.
Schneider line 2 or SL-2 cells of D. melanogaster (
), obtained from Invitrogen (Carlsbad, CA), were used for
promoter assays. Cells were maintained at 25 °C in Schneider's
Drosophila medium (Gibco-BRL) supplemented with 10% heat-
inactivated bovine calf serum and 0.01% penicillin
–streptomy-
cin (Sigma). Every fourth day the cells were transferred to fresh
media.
2.2. Construction of reporter plasmids for Cyp6a2 and Cyp6a8
promoter assay in SL-2 cells
Recombinant plasmids carrying 0.9-kb (
−983/−1) and 0.8-
kb (
−761/−11) upstream DNA of Cyp6a2-91R and Cyp6a8-91R
alleles of 91-R strain respectively (
) were used as templates to amplify different
Table 1
Sequences of the distal primers and PCR conditions used to amplify different regions of the Cyp6a2 and Cyp6a8 genes
Genes
Primer names
Regions amplified
Primer sequences and regions they are identical to
a
T
m
(°C)
Cyp6a2
b
A2-C
−983/−1
5
′-ctcacgcgtTTCATTCGTTTTATCGCCG-3′ (−983/965)
61
A2R-1M
−522/−1
5
′-ccacgcgtCAAGTGGGATCGTCCTGTAC-3′ (−522/−503)
53
A2R-2M
−265/−1
5
′-ccacgcgtGCAGTGAAGCGTATGAGTATC-3′ (−265/−245)
52
A2R-3M
−129/−1
5
′-ccacgcgtGCTAGCTAGCTCACATGCTGTC-3′ (−129/−112)
56
Cyp6a8
c
A8-5F
−491/−11
5
′-ggcctcgagGTTGTGGTAGGTTAGTAGC-3′ (−491/−473)
72.1
A8-1F
−109/−11
5
′-ggcctcgagCTAGCGACTGAGAATGCATC-3′ (−109/−90)
78
A8-0.06F
−67/−11
5
′-ggcctcgagCAGCGTTTAAAAGCAGTTTGC-3′ (−67/−47)
78.6
a
Extra bases added to each primer are shown in lower case. Bases of the upstream DNA are in upper case. For each gene ATG is at + 1. The MluI and XhoI sites
engineered respectively for Cyp6a2 and Cyp6a8 are underlined.
b
The same A2A proximal primer (5
′-ctcgtcgacTTTGCGTAGCTGCTCCC-3′), complimentary to −1/−17 bases of the Cyp6a2 gene, was used with all distal
primers. Engineered bases are in lower case. Added SalI site is underlined. ATG is at + 1. T
m
= 63 °C.
c
G10 proximal primer (5
′-cgttcgaaATGATCTTCGAATACG-3′), complimentary to −11/−26 bases of the Cyp6a8 gene, was used with all distal primers.
Engineered bases are in lower case. The added HindIII site is underlined. ATG is at + 1. T
m
= 70 °C.
57
S. Bhaskara et al. / Gene 377 (2006) 56
–64

regions of the upstream DNA of Cyp6a2 and Cyp6a8 genes via
PCR.
shows the sequences of the distal and proximal
primers used for each Cyp6 gene. Appropriate restriction
enzyme sites that were added to these primers for cloning
purpose are detailed in
. PCR amplifications and
purification of the amplified DNAs were done by using Easy-
Start PCR kit (Molecular Bioscience, Boulder) and kits from
Qiagen (CA), respectively. Four amplified DNA fragments
representing the
−983/−1, −522/−1, −265/−1 and −129/−1
regions (ATG at + 1) of the Cyp6a2 gene were digested with MluI
and SalI, and cloned into MluI
–XhoI cut pGL3-basic vector
(Promega, WI). The resulting recombinant promoter-reporter
plasmids were named 0.98luc-A2, 0.52luc-A2, 0.26luc-A2 and
0.12luc-A2, respectively. In case of Cyp6a8 gene, three
amplified upstream DNAs, representing
−491/−11, −109/−11
and
−67/−11 regions (ATG at +1) of Cyp6a8 were digested with
XhoI and HindIII and cloned into XhoI
–HindIII cut pGL2(N)
vector which is the pGL2-basic vector (Promega, WI) with SalI
site modified to NotI (
). The recombinant
promoter-reporter plasmids were named 0.5luc-A8, 0.1luc-A8
and 0.06luc-A8, respectively. In addition to these plasmids,
0.8luc-A8 and 0.2luc-A8 plasmids carrying
−766/−11 and
−199/−11 upstream DNA of Cyp6a8 (
) were
also used in the present study. All PCR products were sequenced
and found to match with the reported sequence of Cyp6a2 and
Cyp6a8 upstream DNAs (
Dombrowski et al., 1998; Maitra et al.,
).
2.3. Treatment of flies, fly extract preparation, and luciferase assay
Transgenic female flies, 2
–4 days old, were allowed to
feed for 24 h on instant Drosophila food made with aqueous
solution of 2.0 mM Vivarin caffeine (obtained from local
store) or 16 mM pure caffeine (Sigma, St. Louis), a dose
found to be optimum for induction. Food made with water
was used as a control. After treatments, fly extracts were
prepared from groups of ten females for luciferase and
protein assays. Details of fly handling, extract preparation,
firefly luciferase and protein assays have been described
previously (
). Luciferase activity and total
protein in the fly extract were determined by using a
luciferase assay reagent kit (Promega, Madison, WI) and
BCA protein assay kit (Pierce), respectively. Luciferase
activity in fly extracts was expressed as average Registered
Light Units per
μg of total protein (RLU/μg). For each
treatment, extracts prepared from three independent groups of
flies were assayed.
2.4. SL-2 cell transfection, extract preparation and dual
luciferase assay
For transient transfection of SL-2 cells, 1.0
μg of Cyp6a2
or Cyp6a8 firefly luciferase (F-luc) reporter plasmid and
0.5
μg of internal control Renilla luciferase (R-luc) plasmid
(Promega, Madison) were dissolved in sterile 0.1 M CaCl
2
(Invitrogen, CA) and mixed with equal volume of 2× HEPES
buffer (Invitrogen, CA) at pH 7.1 to make calcium phosphate-
DNA mix. Cells to be transfected were resuspended in fresh
SL-2 medium at 1 × 10
6
cells/ml density. To each well of a
48-well microtiter plate, 500
μl of cells were dispensed and to
each well, 84
μl of calcium phosphate-DNA mix was added
in a drop-wise manner. After 18 h incubation at 25 °C, the
medium was replaced with 500
μl of fresh medium and
incubated at 25 °C for 24 h. The cells were then spun down
and resuspended in 500
μl of fresh SL-2 medium containing
8 mM caffeine (Sigma, St. Louis). Following 24 h incubation
at 25 °C, cells were pelleted and 250
μl 1× passive lysis
buffer (Promega, WI) was added to each well. The plates
were agitated on a plate shaker for 15 min at room
temperature to lyse the cells and then centrifuged at
1500 rpm for 5 min at 4 °C. The clear supernatant was
collected from each well and assayed immediately for F-luc
and R-luc activities by using a dual luciferase assay kit
(Promega, WI). Briefly, to 20
μl cell lysate, 100 μl luciferase
assay reagent II was mixed, and placed in a luminometer
within 10 s to measure the firefly luciferase (F-luc) activity.
Immediately after recording the F-luc activity, 100
μl Stop
and Glo reagent (Promega, WI) was added, vortexed gently
and the Renilla luciferase (R-luc) activity was measured
within 10 s. R-luc activity was used to normalize the data,
which were expressed as the ratio of F-luc to R-luc activity.
In each experiment, all reporter constructs of both Cyp6 genes
were used to transfect the same batch of cells. For each
reporter construct, triplicate transfection was done. Two such
experiments were done with two different isolates of each
reporter plasmid DNA to obtain the final mean F-luc/R-luc
ratio.
2.5. Northern blot hybridization
The details of RNA blotting, probe synthesis and
hybridization have been described previously (
). Briefly, total RNA was isolated from caffeine-treated
and untreated adult female flies by using Tri-reagent (Sigma,
MO). For analysis, 20
μg of each RNA sample was dried,
dissolved in 20
μl formaldehyde loading dye solution
(Ambion, Austin, TX) and denatured by incubating at
65 °C for 15 min. Samples were electrophoresed on a
2.2 M formaldehyde
−1.2% agarose gel and blotted onto
nylon membrane. The blots were cut into two pieces to probe
CYP RNA in the upper half and the control RP49 mRNA in
the lower half as described (
). Upper
halves of three blots made from three independent RNA
samples and lower halves of these blots were prehybridized
in separate bottles for 2 h at 37 °C in Northern Max
Prehybridization/Hybridization buffer (Ambion, TX). After
prehybridization,
32
P-labeled CYP (5 × 10
9
cpm/
μg) and RP49
(1.2 × 10
8
cpm/
μg) cDNA probes were added to the buffer in
respective bottles and hybridization was done at 37 °C for
22
–24 h. The blots were washed under high stringent
conditions and scanned with a Packard Radioanalytical
Imager as described (
). Data were
normalized using RP49 hybridization signal as the internal
control.
58
S. Bhaskara et al. / Gene 377 (2006) 56
–64
2.6. Statistics
Data were analyzed using ANOVA program.
3. Results
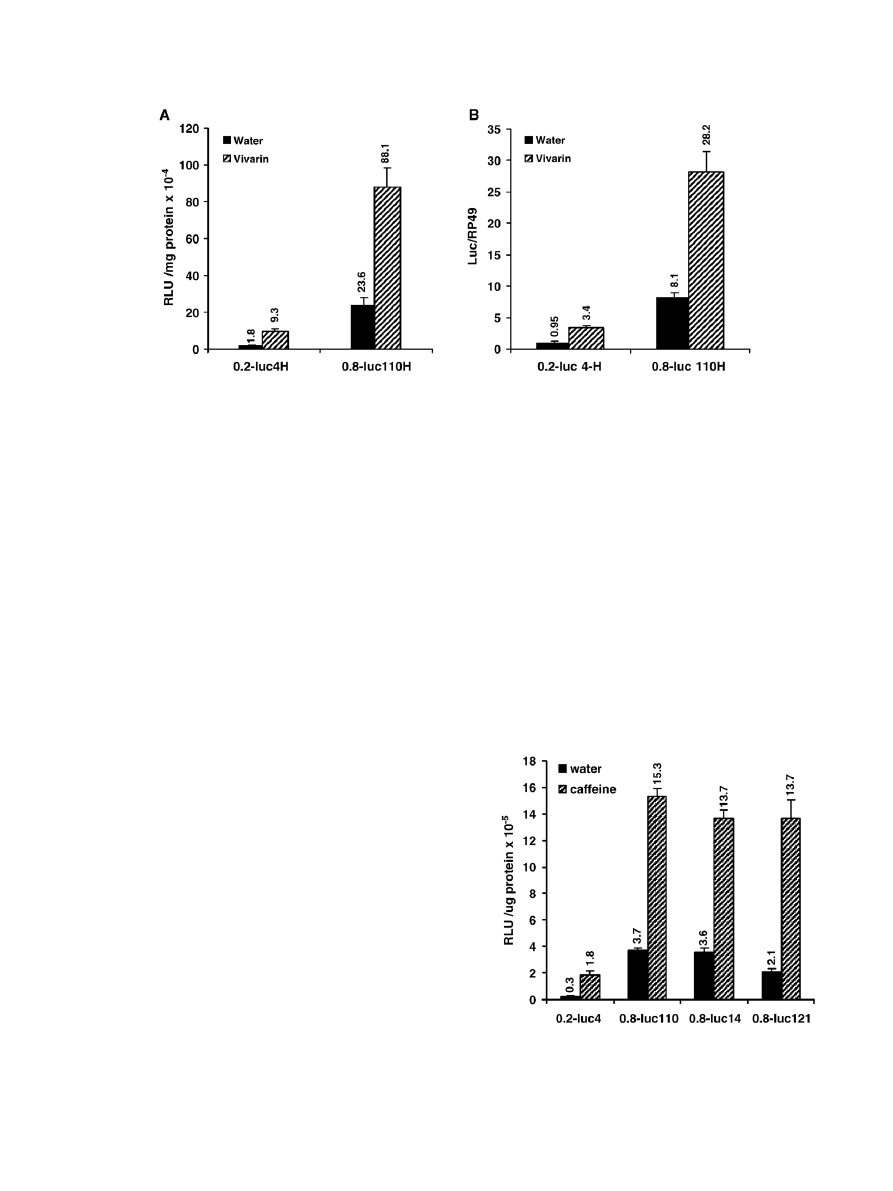
3.1. Vivarin and caffeine induce Cyp6a8 promoter activity
To examine whether caffeine would induce Drosophila
Cyp6a8 gene, adult females of 0.2-luc4H and 0.8-luc110H
transgenic lines were exposed to instant Drosophila medium
made in aqueous solution of over-the-counter caffeine tablet
called Vivarin. The final concentration of caffeine in the me-
dium was 2.0 mM. The results (
A) showed that in both
transgenic lines, luciferase activity was significantly higher in
Vivarin-treated flies compared to the water-treated controls.
Expression of the luc reporter gene was also examined by
Northern blot analysis. A 3-fold induction of LUC mRNA was
observed in both transgenic lines following caffeine treatment
(
B), suggesting that Cyp6a8 promoter is induced by
caffeine. Results also showed that LUC enzyme activity and
LUC mRNA levels were significantly higher in the 0.8-luc line
compared to the 0.2-luc line. These data agree with our previous
observations that 0.8-kb upstream DNA of Cyp6a8 has higher
promoter activity than the 0.2-kb DNA (
Vivarin tablet contains many other ingredients besides
caffeine. Therefore, to examine whether the effect of Vivarin
could be mimicked by caffeine, 0.2-luc4H, 0.8-luc110H, 0.8-
luc14 and 0.8-luc121 transgenic lines were treated with pure
caffeine. Since the latter two strains were not made homozygous
for the reporter transgene, all transgenic lines were crossed to the
ry
506
host strain used for germ line transformation, and the F1
ry
+
(transgene marker) females, heterozygous for the transgene,
were selected and treated with 16 mM caffeine, which was found
to be an optimum concentration in dose response experiments
(data not shown). While a six-fold induction was observed in
0.2-luc4 line, in three 0.8-luc lines the level of induction ranged
from four- to six-fold (
). In another transgenic line (0.2-
luc7), the 0.2-luc transgene located at different chromosomal
location (
) also showed about 5-fold induction
when treated with caffeine (data not shown). Since different
transgenic lines carrying reporter transgene at different chro-
mosomal position showed similar levels of induction, it may be
concluded that caffeine induction is not a result of chromosomal
position effect; it is the function of the Cyp6a8 upstream DNA
driving the luciferase reporter gene.
3.2. Caffeine induces the endogenous Cyp6a8 and Cyp6a2
genes in transgenic and wild type strains
To rule out the possibility that induction of the luc reporter
gene by caffeine is not due to some peculiarity of the reporter
Fig. 1. Expression of luciferase reporter gene in 0.2-luc4H and 0.8-luc110H reporter transgenic lines. Adult females of each line were allowed to feed on
medium containing water or 2.0 mM caffeine (Vivarin) for 24 h. Extracts and RNA prepared from these flies were assayed for luciferase activity (A) and LUC
mRNA level (B) as described in the Materials and methods. The results represent the means ± S.D. of three independent experiments. ANOVA p
b0.0001 for A
and B.
Fig. 2. Effect of pure caffeine on the expression of luciferase reporter transgene
in different transgenic lines. Each transgenic line was crossed to the ry
506
host
strain, and the ry
+
(red-eyed) F1 females carrying the reporter transgene were
allowed to feed on water or 16 mM caffeine containing medium for 24 h.
Extracts prepared from these flies were assayed for luciferase activity. Data
represent mean ± S.D. of three independent determinations. ANOVA p
b0.0001.
59
S. Bhaskara et al. / Gene 377 (2006) 56
–64
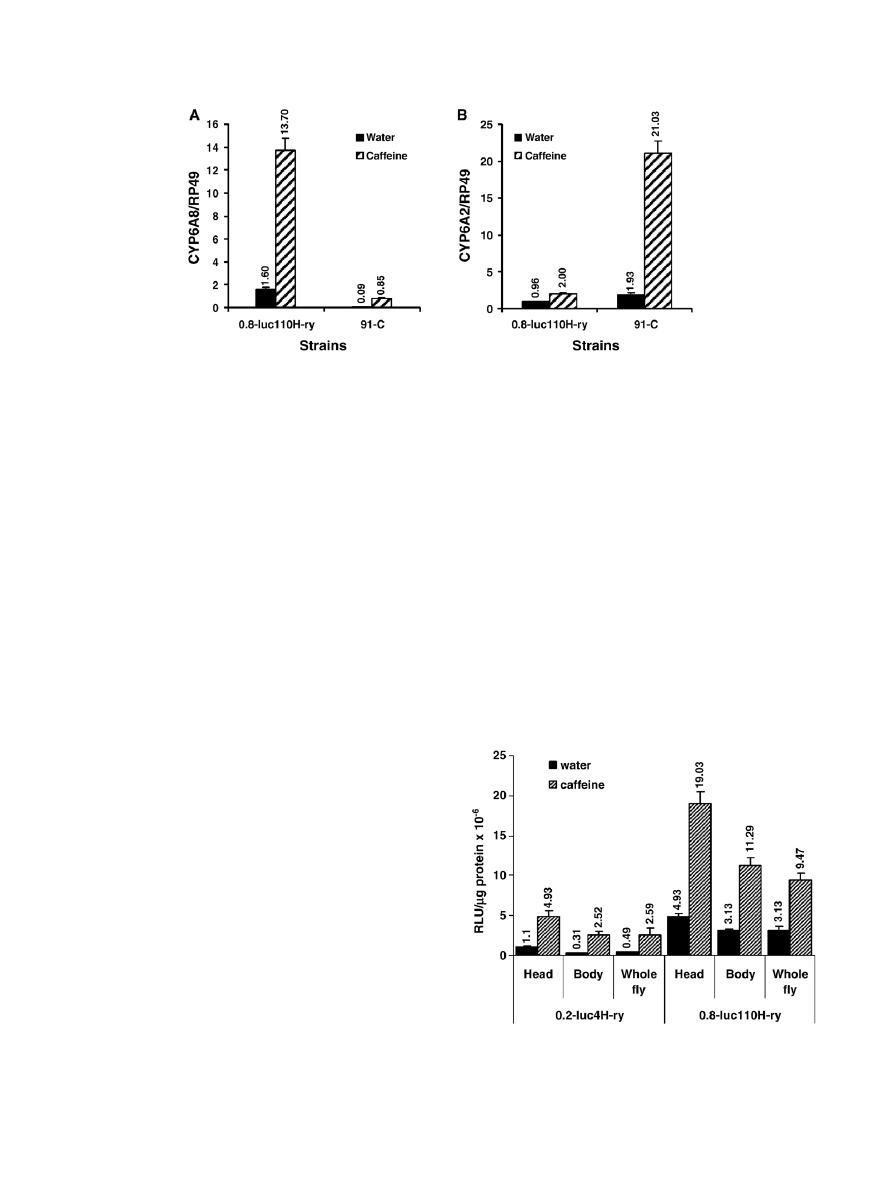
transgenes or the host ry
506
strain, RNA isolated from caffeine-
treated and untreated 0.8-luc110 (H-ry) transgenic flies
homozygous for the reporter transgene and a wild type strain
named 91-C were examined for endogenous Cyp6a8 gene
expression by Northern blot analysis. We also examined
Cyp6a2, another Cyp6 gene of Drosophila that appears to be
under the same gene regulatory mechanism that controls the
Cyp6a8 gene (
). Results showed that in both
0.8-luc110 (H-ry) transgenic line and 91-C strain endogenous
Cyp6a8 and Cyp6a2 genes were induced by caffeine at the
steady-state mRNA level, although the level of induction of
these two genes varied between the two strains (
).
3.3. The 0.2- and 0.8-kb upstream DNAs of Cyp6a8 show
differential promoter activity in different tissues
To investigate the promoter activities of 0.2- and 0.8-kb
upstream DNAs of Cyp6a8 in different parts of the body,
luciferase activity in head, body (whole abdomen plus thorax)
and whole female transgenic flies were measured. Differential
expression of both transgenes was observed in the head and body
of the transgenic flies. Firstly, activities of both 0.2luc-A8 and
0.8luc-A8 transgenes were much higher in the heads than in the
bodies of the untreated control flies (
). Secondly, in the
heads and bodies of the control flies, activity of the 0.8luc-A8
transgene was much higher than the activity of the 0.2luc-A8
transgene. Thirdly, both transgenes were induced by caffeine, but
the level of induced LUC activity was consistently higher in the
0.8-luc110 (H-ry) transgenic line than in the 0.2-luc4 (H-ry) line.
Expression of the 0.2luc-A8 and 0.8luc-A8 transgenes was
also examined in the ovary (
A), gut (
B), abdominal
cuticle plus fat body (
A) and malpighian tubules (
of caffeine-treated and untreated adult female flies. In both
transgenic lines, expression of the reporter gene was very low in
the ovaries of the untreated flies (
A) and caffeine treatment
did not induce the reporter gene in the 0.2-luc4 (H-ry) line
(p = 0.34, Student's t-test). However, in the 0.8-luc110 (H-ry)
line about a 12-fold induction of the reporter gene was observed
in caffeine-treated flies (
A). In the gut (
B), con-
stitutive expression of the reporter transgenes in the untreated
transgenic flies was very low; it was more than one order of
magnitude lower than that found in the ovaries. In fact the
specific activities of luciferase enzyme in treated and untreated
flies in the 0.2-luc4 (H-ry) line were too low to be considered as
an expression of the transgene. In the 0.8-luc110 (H-ry) line,
constitutive expression was also very low but, approximately a
10-fold induction was observed in the caffeine-treated flies
(
B). In the cuticle plus fat body (
A) and in the
malpighian tubules (
B) of the untreated flies of both trans-
genic lines, luciferase activities were found to be several orders
magnitude higher than the activities observed in the ovary and
gut (
). Both transgenes also showed a significant induction in
Fig. 3. Northern blot analysis of endogenous Cyp6a8 and Cyp6a2 gene expression in 0.8-luc110H-ry reporter transgenic line and 91-C wild type strains of Drosophila.
Total RNA was extracted from adult females of each strain, which were treated with water or 16 mM caffeine as described in
. RNA samples were fractionated on
agarose gel, blotted and hybridized with Cyp6a8 (A) and Cyp6a2 (B) gene probes. The levels of RP-49 RNA in the RNA samples were used as internal control to
correct RNA loading discrepancy between lanes. The results represent the means ± S.D. of three independent experiments done with three different RNA samples.
ANOVA p
b0.0001 for A and B.
Fig. 4. Expression of the luciferase reporter transgene in different body parts of
caffeine-treated and untreated female flies of 0.2-luc4H-ry and 0.8-luc110H-ry
transgenic lines. Female flies of each transgenic line were fed with 16 mM
caffeine or water for 24 h and luciferase activity was measured as described. The
results shown are the means ± S.D. of three independent experiments. ANOVA
p
b0.0001.
60
S. Bhaskara et al. / Gene 377 (2006) 56
–64
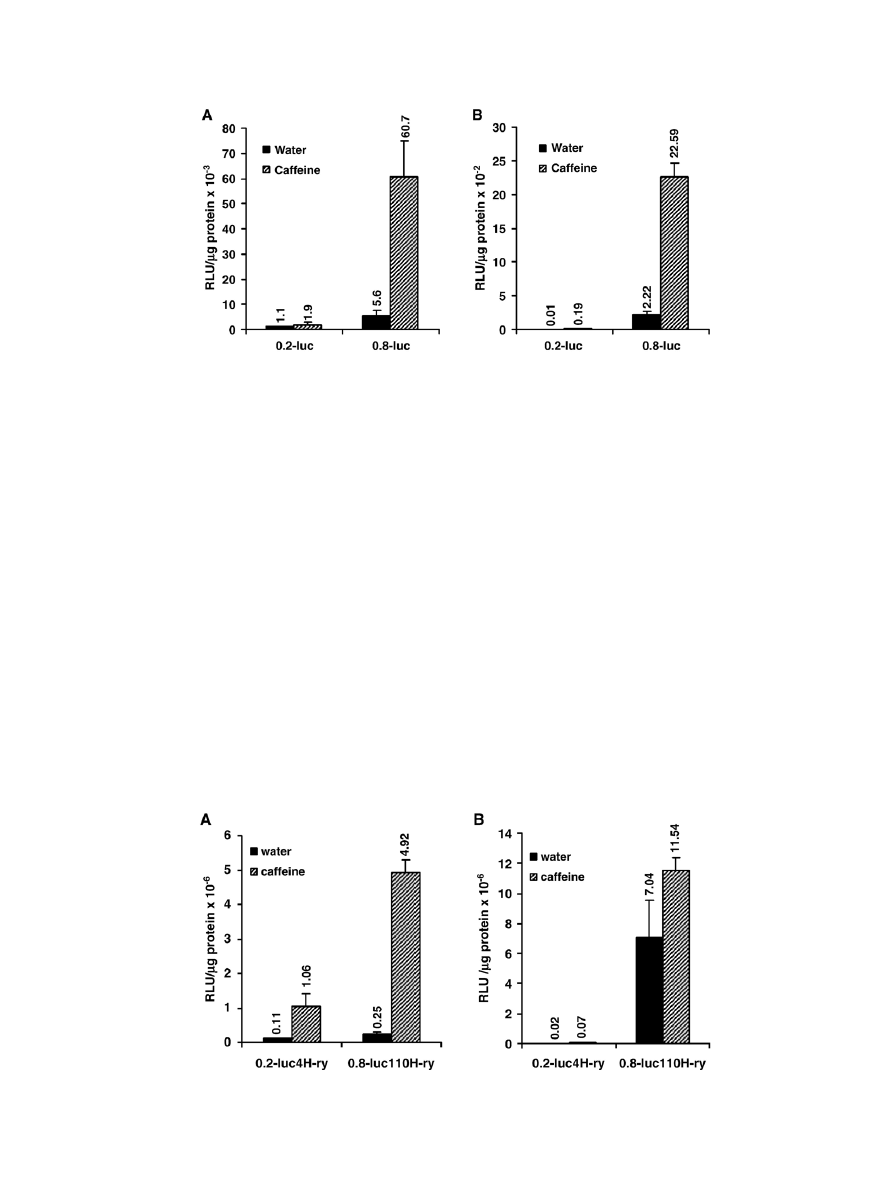
the cuticle plus fat body of the caffeine-treated flies (
A).
However, 0.8-luc110 (H-ry) line showed much higher induction
than the 0.2-luc4 (H-ry) line. Two transgenic lines showed the
most striking difference when the reporter gene activities in the
malpighian tubules of the untreated flies were compared (
B).
In the untreated flies, constitutive expression of the reporter gene
in the 0.8luc-A8 transgenic line was about 350-fold greater than
the expression found in the 0.2luc-A8 line (
B). Although
caffeine treatment induced the reporter gene in both transgenic
lines, the induced activity was still about 160-fold higher in the
0.8-luc110 (H-ry) than that observed in the 0.2-luc4 (H-ry) line.
3.4. Analysis of the Cyp6a2 and Cyp6a8 upstream DNAs for
constitutive and caffeine-induced expression in SL-2 cells
To map the cis-regulatory DNA for constitutive and caffeine-
induced expression, luc reporter plasmids carrying different
lengths of Cyp6a2 or Cyp6a8 upstream DNA were used to
transfect SL-2 cells. The results showed that the basal activity of
0.12luc-A2 and 0.06luc-A8 plasmids carrying the shortest
upstream DNA of Cyp6a2 and Cyp6a8 genes were about 12.6
and 3.3 times greater than the basal activity of the empty pGL3
vector, respectively (
). This high basal activity may be due
to the presence of a putative TATA element in these DNA
fragments (
). When 0.1luc-A8 reporter plas-
mid was used, the basal activity increased about 36% relative to
the activity of the 0.06luc-A8 plasmid and its final activity was
about 4.5-fold higher than the activity of the empty vector
(
B). The basal promoter activity increased another 4- and
5-fold relative to the 0.12luc-A2 and 0.1luc-A8 reporter
plasmids respectively, when 0.26luc-A2 and 0.2luc-A8 plasmids
were used. Compared to the empty vector, these plasmids
showed about a 46- and 21-fold higher basal activity, re-
spectively (
). These data suggest that
−265/−129 Cyp6a2
DNA and
−199/−109 Cyp6a8 have a strong positive effect on
the basal transcription.
In the case of Cyp6a2 gene, increasing the length of the
upstream DNA from
−265 to −522 did not elevate the basal
promoter activity further; instead the promoter activity
decreased by about 42% (
A). Nevertheless, the activity
Fig. 5. Activities of the luciferase reporter gene in the ovary and gut tissues. Female flies of each transgenic line were fed with 16 mM caffeine or water for 24 h.
Ovaries (A) and guts (B) were dissected out from these females and luciferase activity was measured. Means ± S.D. of three independent experiments are shown.
ANOVA p
b0.0001.
Fig. 6. Expression of luciferase reporter gene in fat body and malpighian tubules of adult flies. Female flies of two transgenic lines were treated with 16 mM caffeine or
water, and luciferase activities in the fly extracts were determined as described in Materials and methods. Data shown are the means ± S.D. of three independent
experiments. ANOVA p
b0.0001.
61
S. Bhaskara et al. / Gene 377 (2006) 56
–64
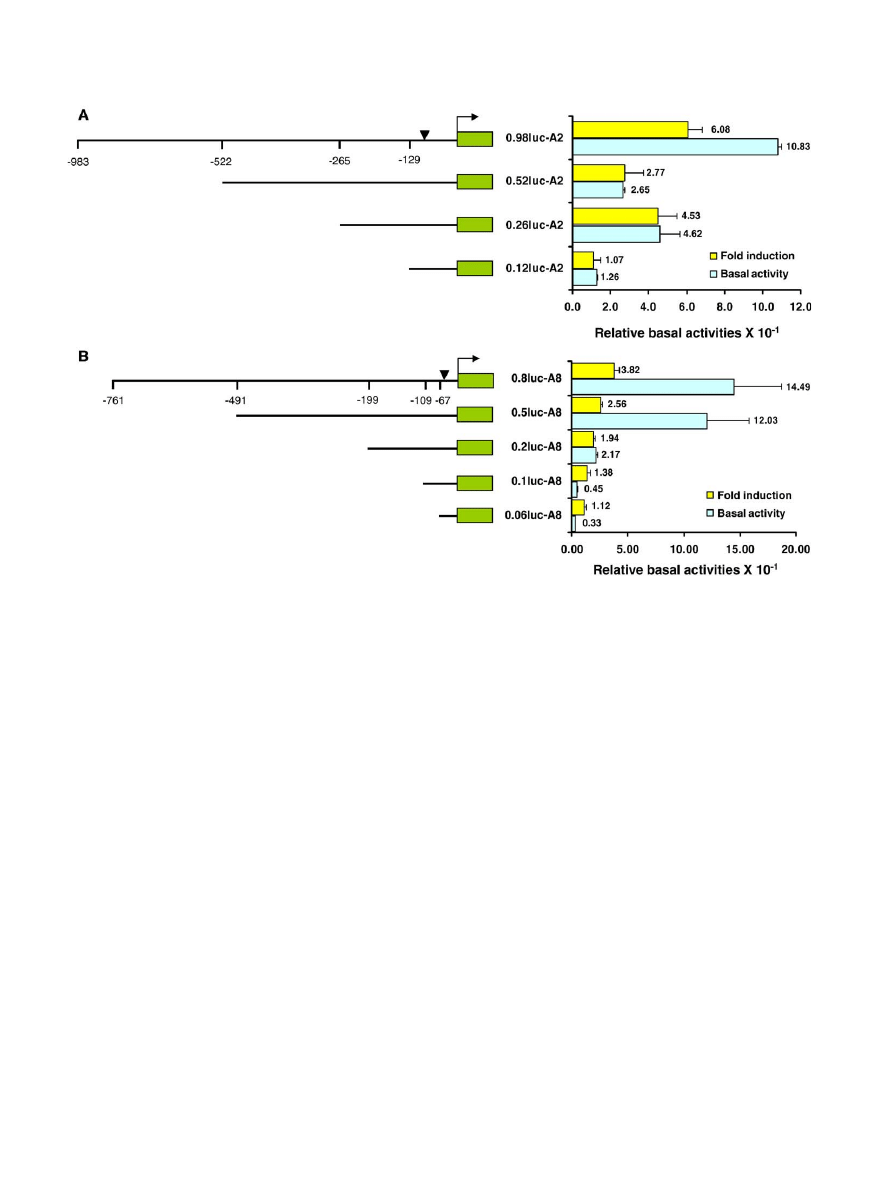
of the 0.52luc-A2 plasmid was still about 26-fold greater than
the activity of the empty vector. Another 4-fold rise in the basal
promoter activity was observed when the upstream DNA length
was increased from
−522 to −983 (
A). Thus, the region
between
−522 and −983 of the Cyp6a2 gene also contains
sequences that have a strong positive effect on the basal
transcription. In the case of Cyp6a8, such sequences appear to
be located between regions
−199 and −491 because the basal
activity of the 0.5luc-A8 reporter plasmid carrying
−491/−11
upstream DNA was about 5.5-fold greater than the activity of
the 0.2luc-A8 reporter plasmid with the shorter (
−199/−11)
upstream DNA.
When transfected SL-2 cells were treated with 8 mM
caffeine, all reporter plasmids, except 0.12luc-A2, 0.1-luc-A8
and 0.06luc-A8, showed significant induction (
). For these
three plasmids, induction level ranged between 1.07- and 1.38-
fold, suggesting that about first 100 bp upstream DNA of both
Cyp6 genes probably lacks caffeine-responsive element.
However, fold-induction increased steadily as the length of
the upstream DNA was increased. The fold-inductions with
0.2luc-A8, 0.5luc-A8 and 0.8luc-A8 reporter plasmids were
1.94, 2.56 and 3.82, respectively (
B). In the case of
Cyp6a2 upstream DNA, the fold-induction profile was a little
different. While the 0.12luc-A2 plasmid did not show much
caffeine induction, a 4.5-fold caffeine induction was observed
with the 0.26luc-A2 plasmid. Similar to the basal activity,
caffeine-induced activity dropped when the 0.52luc-A2 plasmid
was used, but it increased again when the 0.98luc-A2 reporter
plasmid with longer upstream DNA was used. Cells transfected
with this plasmid showed about a 6-fold induction in
transcriptional activity following caffeine treatment, suggesting
that the upstream DNA sequence located between
−522 and
−983 regions plays a strong positive role in caffeine-induced
expression of the Cyp6a2 gene.
4. Discussion
The present investigation clearly demonstrated that the 0.2-
and 0.8-kb upstream DNA of Cyp6a8 show differential
promoter activity in different body parts of adult female D.
melanogaster. The basal promoter activity of both DNA
fragments was found to be highest in the head than in the
body (thorax + abdomen) or the whole fly. When individual
tissues were examined, both DNA fragments showed three and
four orders of magnitude higher promoter activities in the
cuticle plus fat body and malpighian tubules than in the ovaries
Fig. 7. Promoter activities of different upstream DNAs of Cyp6a2 and Cyp6a8 genes in SL-2 cells. SL-2 cells were cotransfected with Renilla luciferase (R-luc) control
plasmid and firefly luciferase (F-luc) chimeric reporter constructs carrying different lengths of upstream DNA of Cyp6a2 or Cyp6a8 gene (ATG at + 1). R-luc and F-luc
activities in the extracts of 8 mM caffeine-treated and untreated transfected cells were determined and compared. Each bar represents mean of two independent
transfection experiments done with two different batches of SL-2 cells and two different isolates of each chimeric reporter plasmid. In each experiment, three aliquots
of cells were transfected with each chimeric reporter plasmid. Relative basal activities were determined by comparing the F-luc/R-luc values of luciferase reporter
plasmid without any promoter and chimeric reporter plasmids. Putative TATA boxes (
▾) of Cyp6a2 and Cyp6a8 are located at −85 and −64 positions, respectively.
ANOVA p
b0.0003 for fold-induction and b0.0001 for basal activity.
62
S. Bhaskara et al. / Gene 377 (2006) 56
–64

and gut, respectively. However, in all tissues, body parts or
whole flies, the 0.8-kb upstream DNA always showed signi-
ficantly higher basal promoter activity than the 0.2-kb upstream
DNA. In the malpighian tubules, the activity of the 0.8-kb DNA
was unusually higher (
∼350-fold) than the activity of the 0.2-kb
DNA. When caffeine-inducibility was compared, both DNA
fragments showed similar fold-induction in head, body and
whole fly. However, in the ovaries, gut and cuticle plus fat body,
0.8-kb DNA showed much higher caffeine-inducibility than the
0.2-kb DNA. Thus, the additional DNA (
−761/−199) present in
the 0.8-kb DNA not only gives a higher basal promoter activity,
but it also gives a higher caffeine-inducibility in ovaries, gut and
cuticle plus fat body. Differential tissue and developmental
expression have also been observed for other Drosophila P450
genes (
Bassett et al., 1997; Maibeche-Coisne et al., 2000
).
In SL-2 cells, the longest upstream DNA of both Cyp6 genes
also showed the highest basal and caffeine-induced promoter
activity. However, basal promoter activity increased in two
steps when the length of the upstream DNA of Cyp6 gene was
increased. In the case of Cyp6a2 gene, sequences giving two-
step increase of promoter activity are located in
−265/−129 and
−983/−522 regions of the upstream DNA. These sequences
also increased caffeine-inducibility. In the case of Cyp6a8 gene,
the
−199/−109 DNA gave a 4-fold and the −491/−199 DNA
gave about a 6-fold rise in basal promoter activity. Caffeine-
inducibility of the Cyp6a8 upstream DNA, however, increased
progressively as its length was increased from position
−109 to
−761. The −109/−11 DNA of Cyp6a8 did not show any
caffeine-inducibility. These observations suggest that sequences
present in the longest upstream of each Cyp6 gene act
synergistically and give the highest basal and caffeine-induced
expression. It is not known whether the same sequences are
involved in basal and caffeine-induced activity.
showed that
−117/−75 region of rat
dopamine 2 receptor (D2R) gene containing putative binding
sites for dopamine receptor regulating factor (DRRF) and SP1
showed a 1.9-fold induction in caffeine-treated cells. It has been
hypothesized that stimulation of the D2R gene by caffeine is
probably mediated by alleviating the repressive activity of DRRF
(
).
have shown that
DRRF, a zinc finger type transcription factor, binds to GT and GC
boxes and displaces the Sp1 and Sp3 transcription factors.
Although Alibaba2.1 program (
com/pub/programs/alibaba2/index.html
) identified putative Sp1
sites at
−944/−934 and −17/−8 positions of Cyp6a2, and at
−736/−737 and −641/−631 positions of Cyp6a8 genes, in SL-2
cells these sites are probably not involved in the expression of
Cyp6a2 and Cyp6a8 genes because Sp1 proteins are not found in
this cell line (
). Therefore, the constitutive
and caffeine-induced expression found in SL-2 cells must be
mediated by factors other than Sp1. However, involvement of the
putative Sp1 sites in the regulation of Cyp6 gene promoter in the
adult flies cannot be ruled out.
In two studies (
Goasduff et al., 1996; Svenningsson et al.,
), caffeine has been shown to increase the steady-state level
of two CYP mRNAs, and RNAs for FOS and JUN family
proteins. The homo- and heterodimer of these proteins are called
activator protein or AP1, which binds with the AP1 element and
enhances promoter activity. The AP1 proteins are products of
immediate early genes, which are induced by various extracel-
lular stimuli via cAMP pathway (
). Many cAMP
responsive gene promoters are known to have AP1 and cAMP
response element or CRE, which binds with CRE binding
protein called CREB or CRE-BP (
and references
therein). Analysis of the Cyp6a2 and Cyp6a8 upstream DNAs
with MATCH
™ program (
) at 0.8 core and matrix
similarities identified 598 transcriptionally important sequence
motifs including many potential AP-1 and CREB sites (
).
Since 0.12luc-A2 and 0.1luc-A8 DNA did not show any
Table 2
Locations of putative sequence motifs in the upstream DNA of Cyp6a2 and
Cyp6a8 that participate in cAMP-mediated transcriptional regulation
Gene
Position
(strand)
Core
match
Matrix
match
Sequence
a
(+ strand)
Factor
name
Cyp6a2
−865 (−)
0.850
0.834
aattaTGTCAtt
CREB
−864 (−)
0.967
0.940
attaTGTCAtt
AP-1
−863 (−)
0.848
0.865
ttATGTCa
CRE-BP1
−741 (−)
0.800
0.804
gCTCCTta
STRE
−648 (−)
0.850
0.834
tcttaTGTCAaa
CREB
−647 (−)
0.967
0.878
cttaTGTCAaa
AP-1
−646 (−)
0.818
0.842
ttaTGTCA
CREB
−549 (+)
0.831
0.891
TTACTtaa
CRE-BP1
−489 (−)
0.955
0.850
tTGAATgac
AP-1
−486 (+)
0.962
0.918
aaTGACCgtgt
AP-1
−482 (−)
0.967
0.927
accgTGTCAtt
AP-1
−371 (+)
0.811
0.802
aaTGAGTgata
AP-1
−370 (−)
0.989
0.880
aTGAGTgat
AP-1
−182 (+)
0.967
0.899
caTGACAacaa
AP-1
−161 (−)
1.000
0.920
gcgtAGTCAtg
AP-1
−120 (−)
0.967
0.896
atgcTGTCAtg
AP-1
−99 (+)
1.000
0.975
gcAGGGGa
STRE
Cyp6a8
−749 (+)
1.000
0.999
taAGGGGg
STRE
−695 (+)
0.818
0.823
TAACGtag
CREB
−524 (+)
0.967
0.945
aaTGACAcact
AP-1
−523 (−)
0.804
0.852
aTGACAcac
AP-1
−510 (−)
1.000
0.919
atagAGTCAtg
AP-1
−441 (−)
0.962
0.889
gtctGGTCAac
AP-1
−390 (+)
0.811
0.813
cgTGAGTaagc
AP-1
−389 (−)
0.989
0.898
gTGAGTaag
AP-1
−328 (−)
0.839
0.839
tgataAGTCAca
CREB
−327 (−)
1.000
0.962
gataAGTCAca
AP-1
−326 (+)
1.000
0.884
ataAGTCAc
AP-1
−318 (−)
1.000
0.927
cagttCGTCAct
CREB
−317 (−)
0.967
0.954
agttCGTCAct
AP-1
−316 (−)
1.000
0.889
gttCGTCActgt
CREB
−292 (−)
0.811
0.807
cattATTCAtc
AP-1
−291 (+)
0.955
0.880
attATTCAt
AP-1
−133 (−)
0.831
0.815
taaAGTAA
CRE-BP1
−95 (−)
1.000
0.849
tgcatCGTCAtg
CREB
−94 (−)
0.967
0.880
gcatCGTCAtg
AP-1
−93 (−)
1.000
0.888
catCGTCAtgca
CREB
−81 (−)
0.839
0.800
aagtaAGTCAat
CREB
−80 (−)
1.000
0.958
agtaAGTCAat
AP-1
−80 (−)
1.000
0.957
agtaAGTCAat
AP-1
−79 (+)
1.000
0.881
gtaAGTCAa
AP-1
−79 (−)
0.818
0.819
gtaAGTCA
CREB
−39 (+)
1.000
0.849
TTACGgaa
CRE-BP1
a
The capital letters indicate the positions in the sequence which match with
the core sequence of the matrix.
63
S. Bhaskara et al. / Gene 377 (2006) 56
–64

caffeine-inducibility (
), the AP-1 and CREB sites found
within about
−100 bp upstream DNA of both genes may not be
involved in caffeine induction. However, the CREB-BP and AP-
1 sites found at
−133 position of the Cyp6a8, and at −161 and
−182 positions of the Cyp6a2 (
) may play a role in
caffeine induction because 0.2luc-A8 and 0.26luc-A2 DNA
carrying these elements gave about 2- and 4.5-fold caffeine
induction, respectively. Similarly, the putative AP-1 and CREB
sites found in the DNA upstream of position
−549 of the Cyp6a2
and
−510 of the Cyp6a8 genes (
) may be important for
caffeine induction because the longest reporter constructs
(0.98luc-A2 and 0.8luc-A8) carrying these putative AP1 and
CREB sites gave further increase in fold-induction (
).
Cyp6a2 and Cyp6a8 DNA upstream of positions
−549 and
−510, respectively, also have a sequence that matches with the
stress response element (STRE) found in many yeast genes
(
). Since caffeine is thought to cause cellular
stress, the putative STREs may also play a role in caffeine
induction of the Cyp6a2 and Cyp6a8 genes. Refined mapping
coupled with site-directed mutagenesis will be necessary to
determine the role of putative AP-1, CREB and STRE elements.
If AP-1 and/or CREB are indeed involved in caffeine-induced
transcription, it may be hypothesized that caffeine-induction of
Cyp6 genes is mediated via cAMP signaling pathway. This
hypothesis is currently being investigated.
Acknowledgements
The project was supported by the National Research
Initiative of the USDA Cooperative State Research, Education
and Extension Service, grant number 2002-35302-12281 to RG.
References
Bassett, M.H., McCarthy, J.L., Waterman, M.R., Sliter, T.J., 1997. Sequence and
developmental expression of Cyp18, a member of a new cytochrome P450
family from Drosophila. Mol. Cell. Endocrinol. 131, 39
–49.
Brandt, A., et al., 2002. Differential expression and induction of two Drosophila
cytochrome P450 genes near the Rst(2)DDT locus. Insect Mol. Biol. 11,
337
–341.
Courey, A.J., Tjian, R., 1988. Analysis of Sp1 in vivo reveals multiple
transcriptional domains, including a novel glutamine-rich activation motif.
Cell 55, 887
–898.
Daborn, P., Boundy, S., Yen, J., Pittendrigh, B., ffrench-Constant, R., 2001.
DDT resistance in Drosophila correlates with Cyp6g1 over-expression and
confers cross-resistance to the neonicotinoid imidacloprid. Mol. Genet.
Genomics 266, 556
–563.
Dombrowski, S.M., et al., 1998. Constitutive and barbital-induced expression of
the Cyp6a2 allele of a high producer strain of CYP6A2 in the genetic
background of a low producer strain. Gene 221, 69
–77.
Fredholm, B.B., Battig, K., Holmen, J., Nehlig, A., Zvartau, E.E., 1999. Actions
of caffeine in the brain with special reference to factors that contribute to its
widespread use. Pharmacol. Rev. 51, 83
–133.
Goasduff, T., Dreano, Y., Guillois, B., Menez, J.F., Berthou, F., 1996. Induction
of liver and kidney CYP1A1/1A2 by caffeine in rat. Biochem. Pharmacol.
52, 1915
–1919.
Guengerich, F.P., 2004. Cytochrome P450: what have we learned and what are
the future issues? Drug Metab. Rev. 36, 159
–197.
Hankinson, O., 2005. Role of coactivators in transcriptional activation by the
aryl hydrocarbon receptor. Arch. Biochem. Biophys. 433, 379
–386.
Hwang, C.K., et al., 2001. Dopamine receptor regulating factor, DRRF: a zinc
finger transcription factor. Proc. Natl. Acad. Sci. U. S. A. 98, 7558
–7563.
Kel, A.E., Gossling, E., Reuter, I., Cheremushkin, E., Kel-Margoulis, O.V.,
Wingender, E., 2003. MATCH: a tool for searching transcription factor
binding sites in DNA sequences. Nucleic Acids Res. 31, 3576
–3579.
Lorist, M.M., Tops, M., 2003. Caffeine, fatigue, and cognition. Brain Cogn. 53,
82
–94.
Maibeche-Coisne, M., Monti-Dedieu, L., Aragon, S., Dauphin-Villemant, C.,
2000. A new cytochrome P450 from Drosophila melanogaster, CYP4G15,
expressed in the nervous system. Biochem. Biophys. Res. Commun. 273,
1132
–1137.
Maitra, S., Dombrowski, S.M., Waters, L.C., Ganguly, R., 1996. Three second
chromosome-linked clustered Cyp6 genes show differential constitutive and
barbital-induced expression in DDT-resistant and susceptible strains of
Drosophila melanogaster. Gene 180, 165
–171.
Maitra, S., Dombrowski, S.M., Basu, M., Raustol, O., Waters, L.C., Ganguly,
R., 2000. Factors on the third chromosome affect the level of Cyp6a2 and
Cyp6a8 expression in Drosophila melanogaster. Gene 248, 147
–156.
Maitra, S., Price, C., Ganguly, R., 2002. Cyp6a8 of Drosophila melanogaster:
gene structure, and sequence and functional analysis of the upstream DNA.
Insect Biochem. Mol. Biol. 32, 859
–870.
Mandal, P.K., 2005. Dioxin: a review of its environmental effects and its aryl
hydrocarbon receptor biology. J. Comp. Physiol., B 175, 221
–230.
Parrou, J.L., Enjalbert, B., Francois, J., 1999. STRE- and cAMP-independent
transcriptional induction of Saccharomyces cerevisiae GSY2 encoding
glycogen synthase during diauxic growth on glucose.
Sahir, N., Evrard, P., Gressens, P., 2004. Caffeine induces sonic hedgehog gene
expression in cultured astrocytes and neurons. J. Mol. Neurosci. 24, 201
–206.
Schneider, I., 1972. Cell lines derived from late embryonic stages of Drosophila
melanogaster. J. Embryol. Exp. Morphol. 27, 353
–365.
Scott, J.G., 1999. Cytochrome P450 and insecticide resistance. Insect Mol. Biol.
29, 757
–777.
Sng, J.C., Taniura, H., Yoneda, Y., 2004. A tale of early response genes. Biol.
Pharm. Bull. 27, 606
–612.
Stonehouse, A.H., Adachi, M., Walcott, E.C., Jones, F.S., 2003. Caffeine regulates
neuronal expression of the dopamine 2 receptor gene. Mol. Pharmacol. 64,
1463
–1473.
Sueyoshi, T., Negishi, M., 2001. Phenobarbital response elements of
cytochrome P450 genes and nuclear receptors. Annu. Rev. Pharmacol.
Toxicol. 41, 123
–143.
Svenningsson, P., Strom, A., Johansson, B., Fredholm, B.B., 1995. Increased
expression of c-jun, junB, AP-1, and preproenkephalin mRNA in rat striatum
following a single injection of caffeine. J. Neurosci. 15, 3583
–3593.
Wang, H., Negishi, M., 2003. Transcriptional regulation of cytochrome p450 2B
genes by nuclear receptors. Curr. Drug Metab. 4, 515
–525.
64
S. Bhaskara et al. / Gene 377 (2006) 56
–64
Document Outline
- Induction of two cytochrome P450 genes, Cyp6a2 and Cyp6a8, of Drosophila melanogaster by caffei.....
- Introduction
- Materials and methods
- Results
- Vivarin and caffeine induce Cyp6a8 promoter activity
- Caffeine induces the endogenous Cyp6a8 and Cyp6a2 genes in transgenic and wild type strains
- The 0.2- and 0.8-kb upstream DNAs of Cyp6a8 show differential promoter activity in different ti.....
- Analysis of the Cyp6a2 and Cyp6a8 upstream DNAs for constitutive and caffeine-induced expressio.....
- Discussion
- Acknowledgements
- References
Wyszukiwarka
Podobne podstrony:
A comparison of Drosophila melanogaster detoxication gene induction responses for six insecticides,
Correlates of Sleep and Waking in Drosophila melanogaster
William Pelfrey Billy, Alfred, and General Motors, The Story of Two Unique Men, a Legendary Company
Two Trips to Gorilla Land and the Cataracts of the Congo Volume 1 by Richard F Burton
Znaczenie enzymów cytochromu P450
USŁUGI, World exports of commercial services by region and selected economy, 1994-04
Effect of caffeine on fecundity egg laying capacity development time and longevity in Drosophila
ENHANCEMENT OF HIV 1 REPLICATION BY OPIATES AND COCAINE THE CYTOKINE CONNECIOION
Shaman Saiva and Sufi A Study of the Evolution of Malay Magic by R O Winstedt
Dream Yoga and the practice of Natural Light by Namkhai Norbu
Beowulf, Byrhtnoth, and the Judgment of God Trial by Combat in Anglo Saxon England
Every Goodbye Aint Gone An Anthology of Innovative Poetry by African Americans Modern and Contempor
Philosophy Of Mind Minds,Machines,And Mathematics A Review Of Shadows Of The Mind By Roger Penrose
Claus Dieter Meyer & Karsten Muller The Magic of Chess Tactics by Claus Dieter Meyer and Karsten Mu
Replicated triangle and duo–trio tests Discrimination capacity of assessors evaluated by Bayes rule
Psychic Vampire Codex A Manual of Magick and Energy Work Wicca Occult by Michelle A Belanger
Effect of caffeine on fecundity egg laying capacity development time and longevity in Drosophila
Drying kinetics and quality of beetroots dehydrated by combination of convective and vacuum microwav
więcej podobnych podstron