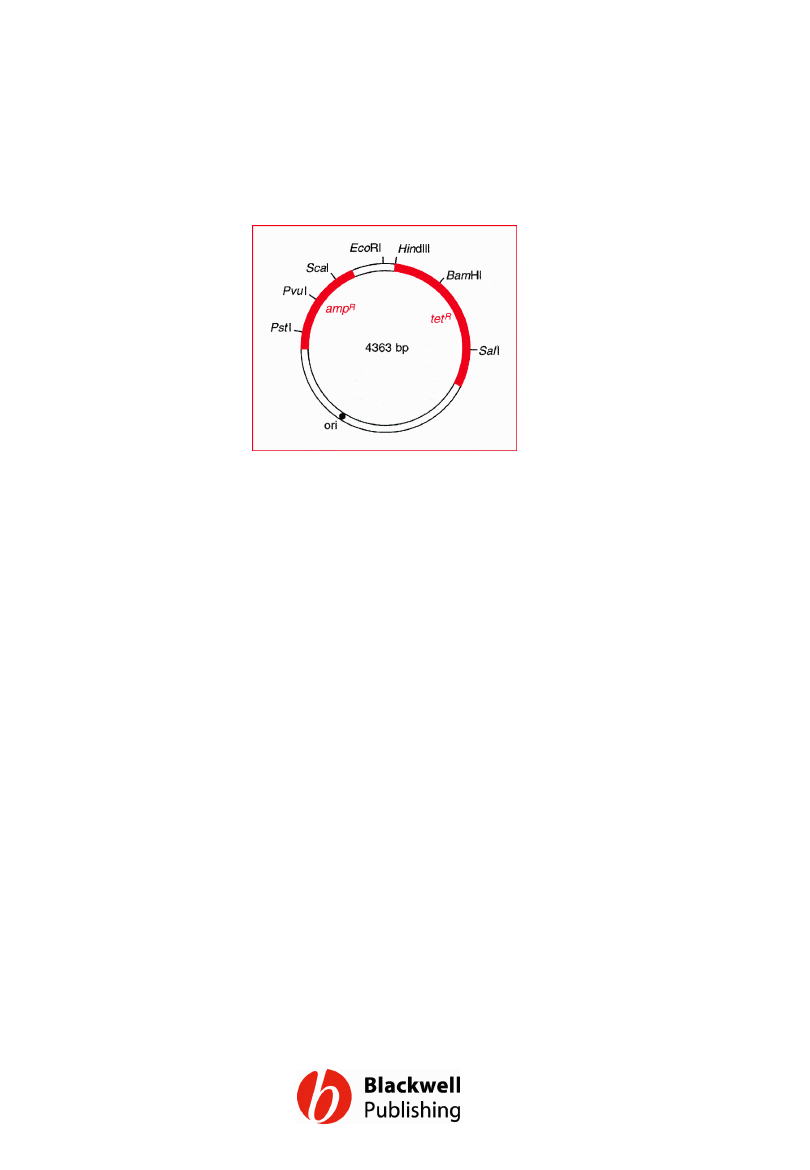
Figure 6.1 A map of pBR322 showing the
positions of the ampicillin resistance (ampR)
and tetracycline resistance (tetR) genes, the
origin of replication (ori) and some of the most
important restriction sites.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
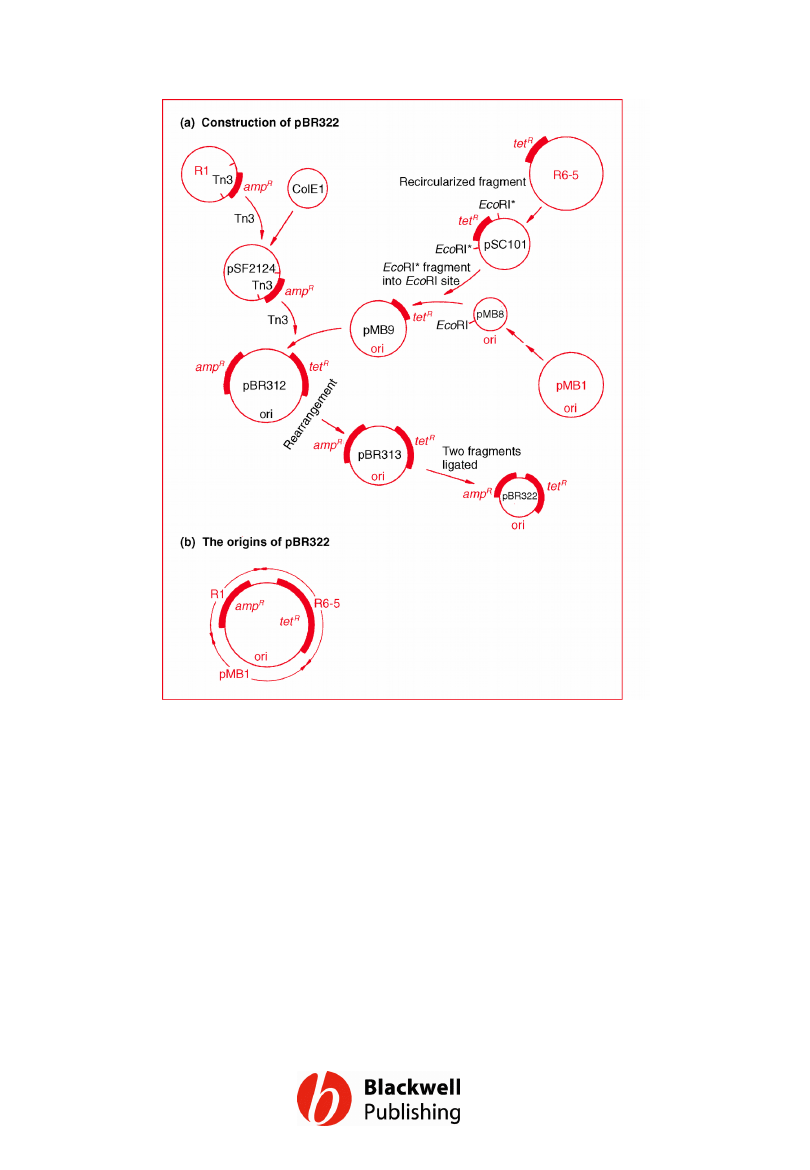
Figure 6.2 The pedigree of pBR322. (a) The
manipulations involved in construction of
pBR322. (b) A summary of the origins of
pBR322.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
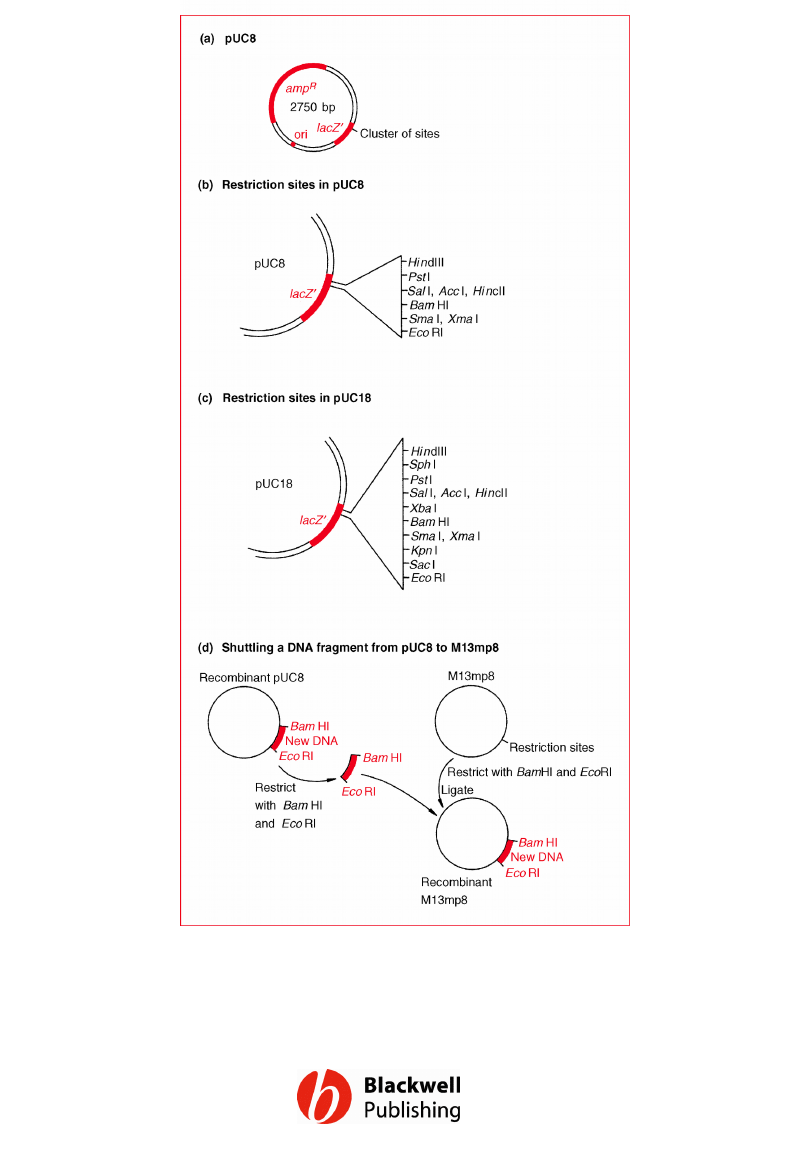
Figure 6.3 The pUC plasmids. (a) The structure of pUC8.
(b) The restriction site cluster in the lacZ¢ gene of pUC8.
(c) The restriction site cluster in pUC18. (d) Shuttling a
DNA fragment from pUC8 to M13mp8.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
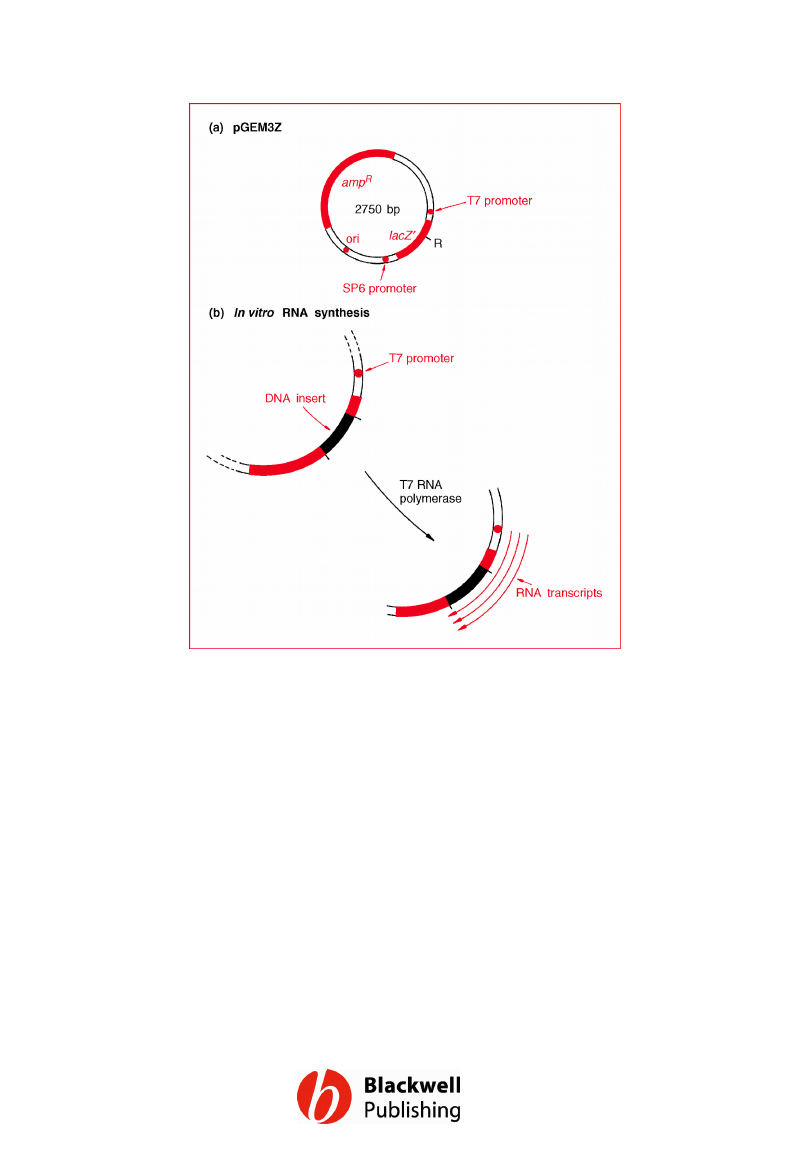
Figure 6.4 pGEM3Z. (a) Map of the vector. (b)
In vitro RNA synthesis. R = cluster of
restriction sites for EcoRI, SacI, KpnI, AvaI,
SmaI, BamHI, XbaI, SalI, AccI, HincII, PstI, SphI
and HindIII.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
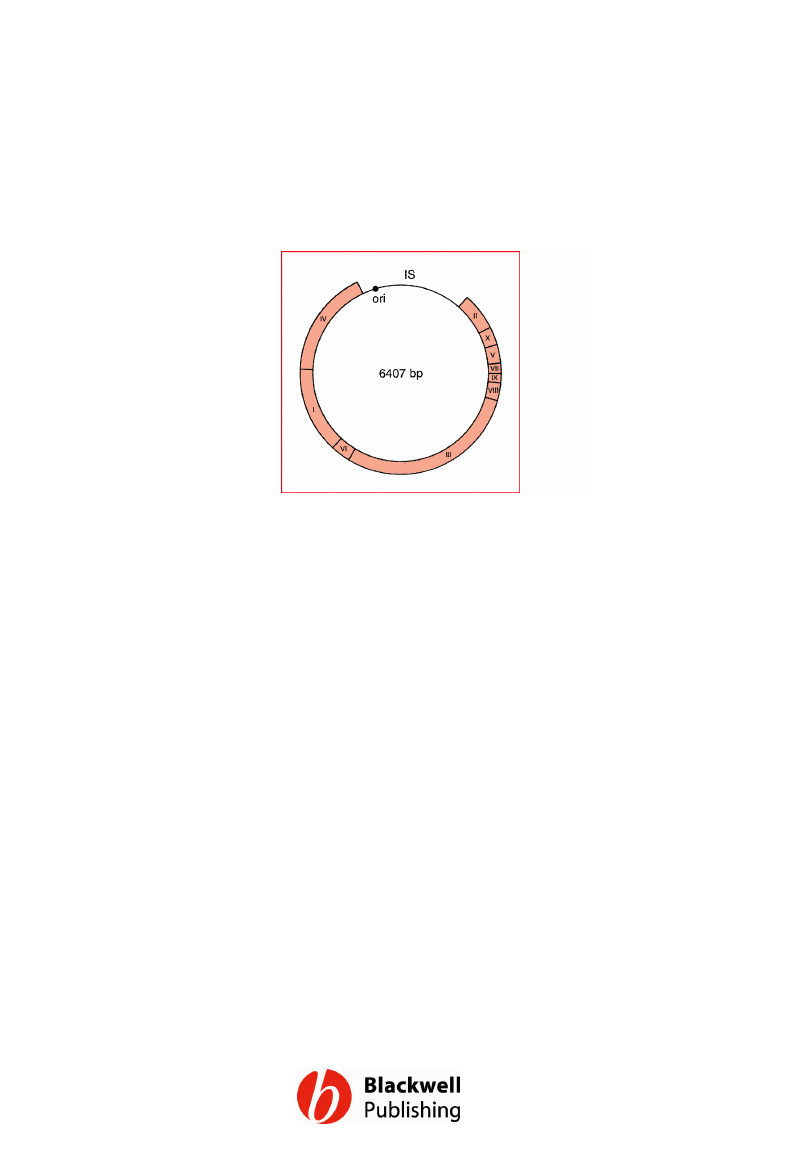
Figure 6.5 The M13 genome, showing the
positions of genes I to X.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
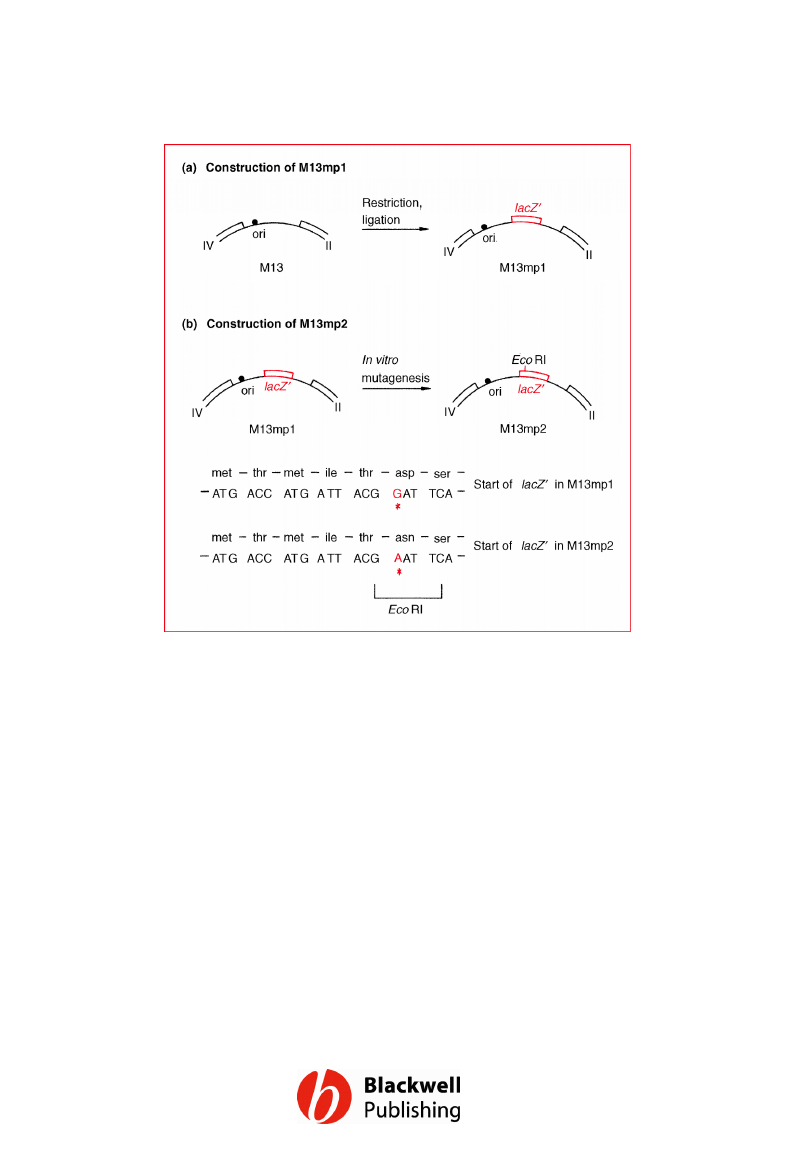
Figure 6.6 Construction of (a) M13mp1, and
(b) M13mp2 from the wild-type M13 genome.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
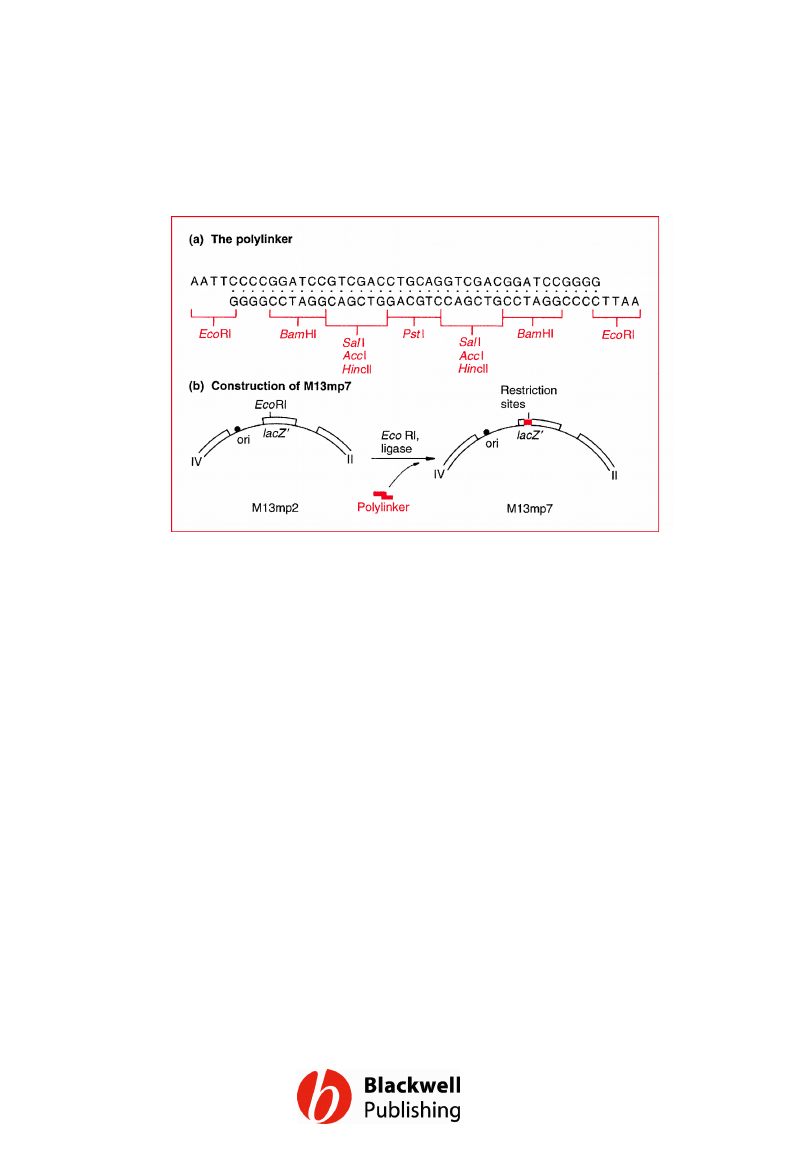
Figure 6.7 Construction of M13mp7: (a) the
polylinker, and (b) its insertion into the EcoRI
site of M13mp2. Note that the SalI restriction
sites are also recognized by AccI and HincII.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
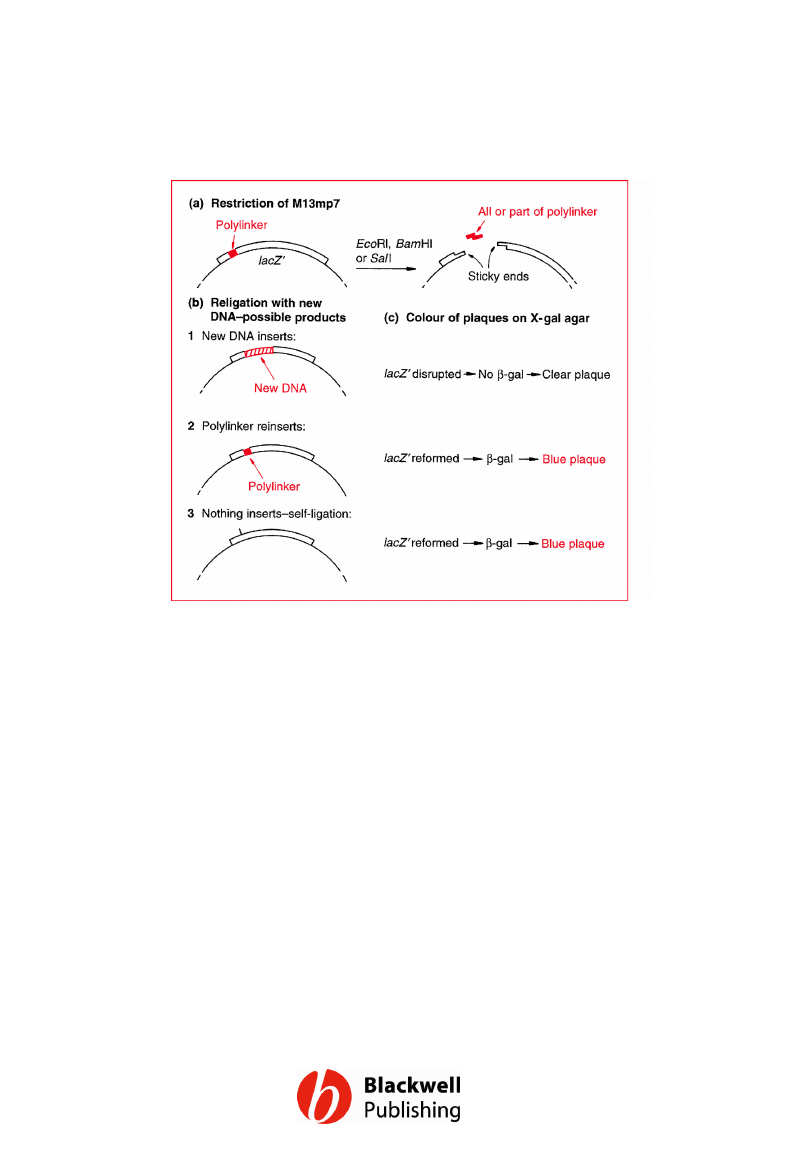
Figure 6.8 Cloning with M13mp7 (see text for
details).
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
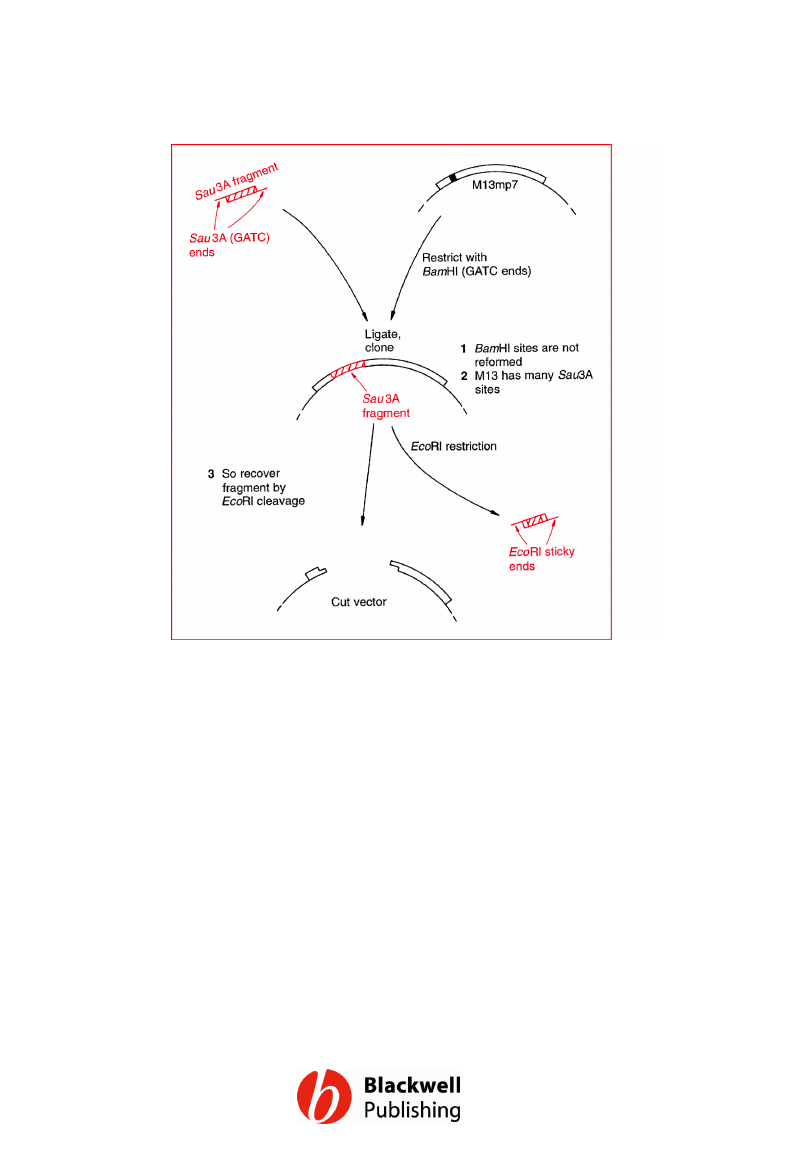
Figure 6.9 Recovery of cloned DNA from a
recombinant M13mp7 molecule by restriction
at the outer sites of the polylinker.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
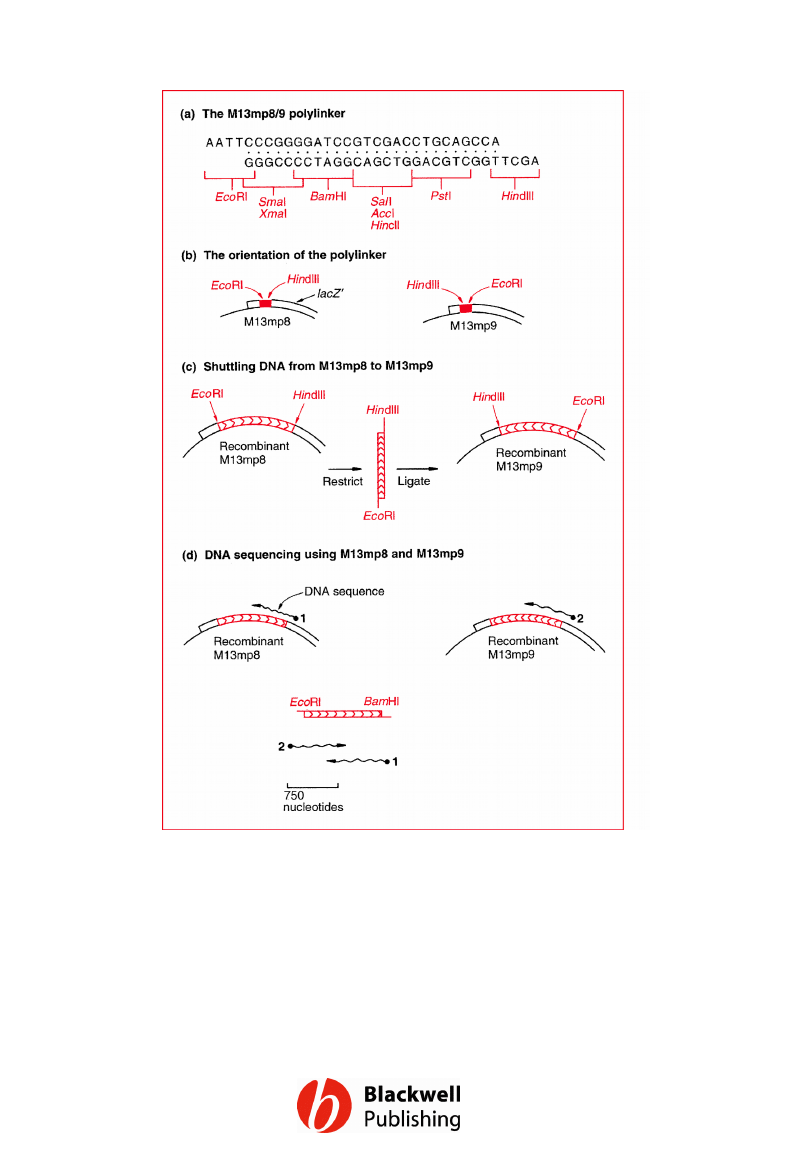
Figure 6.10 M13mp8 and M13mp9.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
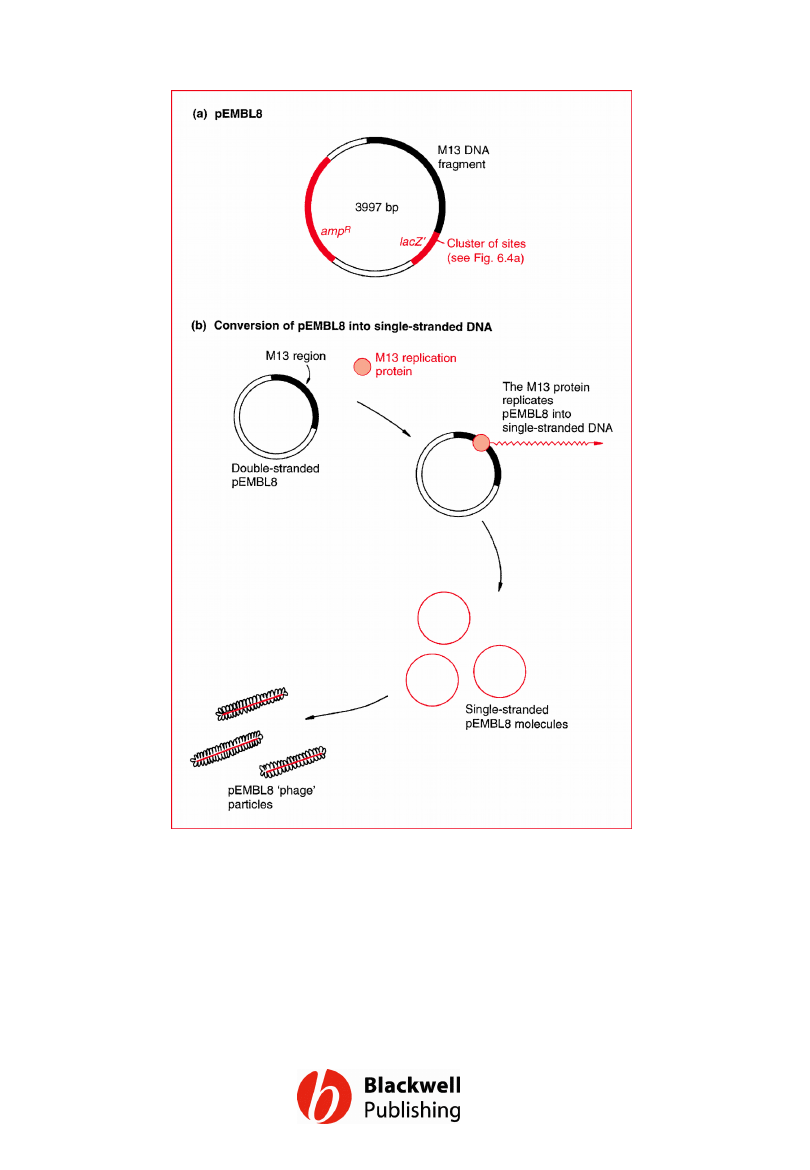
Figure 6.11 pEMBL8: a hybrid plasmid–M13
vector that can be converted into single-
stranded DNA.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
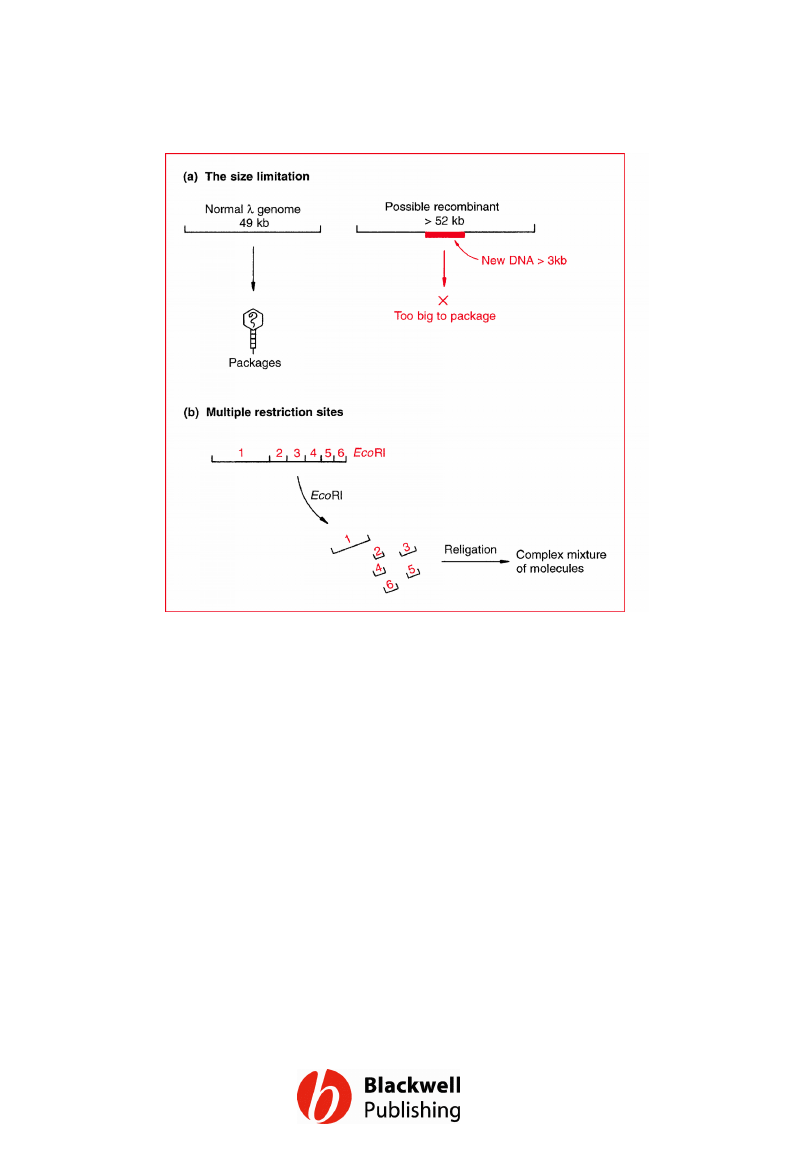
Figure 6.12 The two problems that had to be
solved before l cloning vectors could be
developed. (a) The size limitation placed on
the l genome by the need to package it into
the phage head. (b) l DNA has multiple
recognition sites for almost all restriction
endonucleases.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.

Figure 6.13 The l genetic map, showing the
position of the main non-essential region that
can be deleted without affecting the ability of
the phage to follow the lytic infection cycle.
There are other, much shorter non-essential
regions in other parts of the genome.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
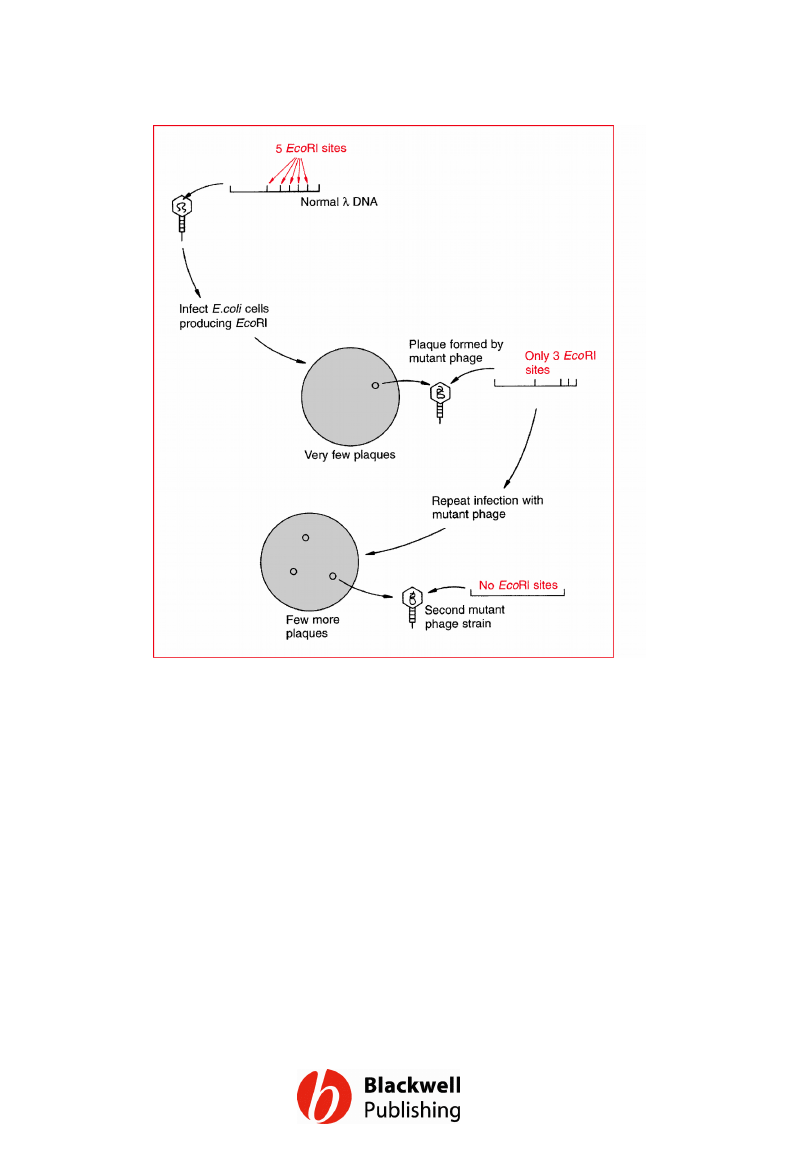
Figure 6.14 Using natural selection to isolate
l phage lacking EcoRI restriction sites.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
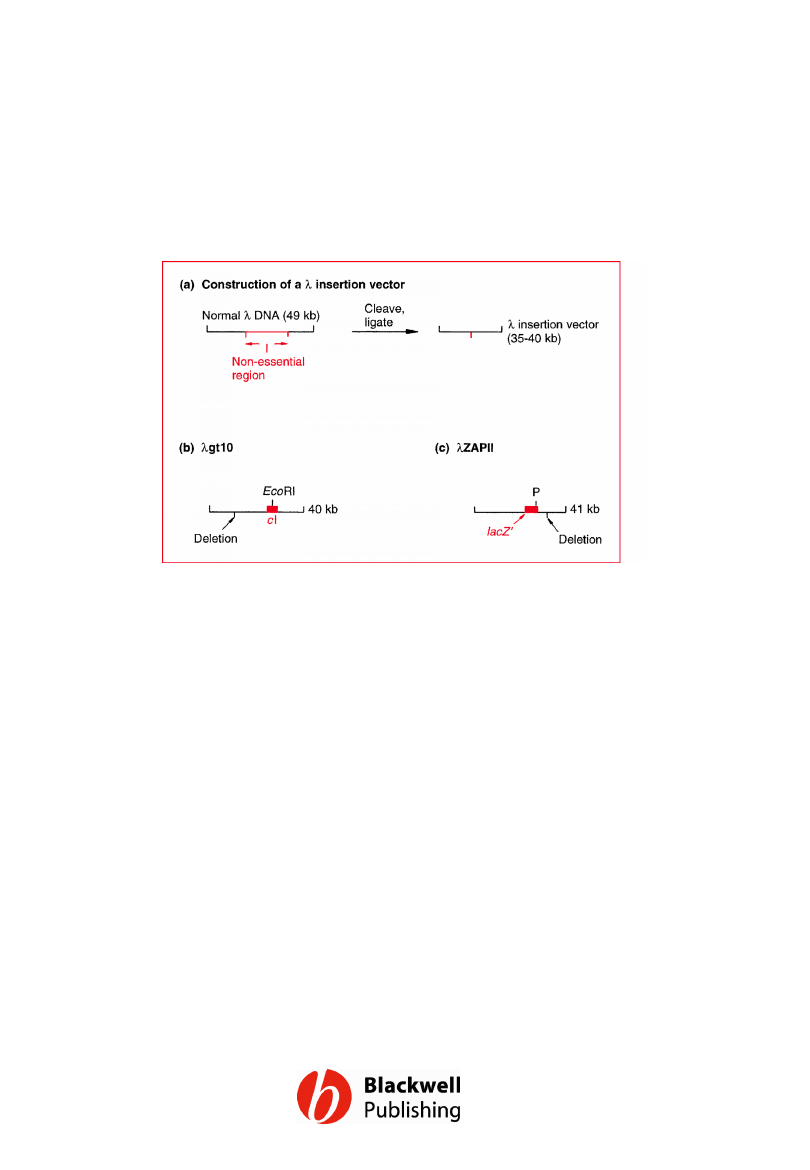
Figure 6.15 l insertion vectors. P = polylinker
in the lacZ¢ gene of lZAPII, containing unique
restriction sites for SacI, NotI, XbaI, SpeI,
EcoRI and XhoI.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
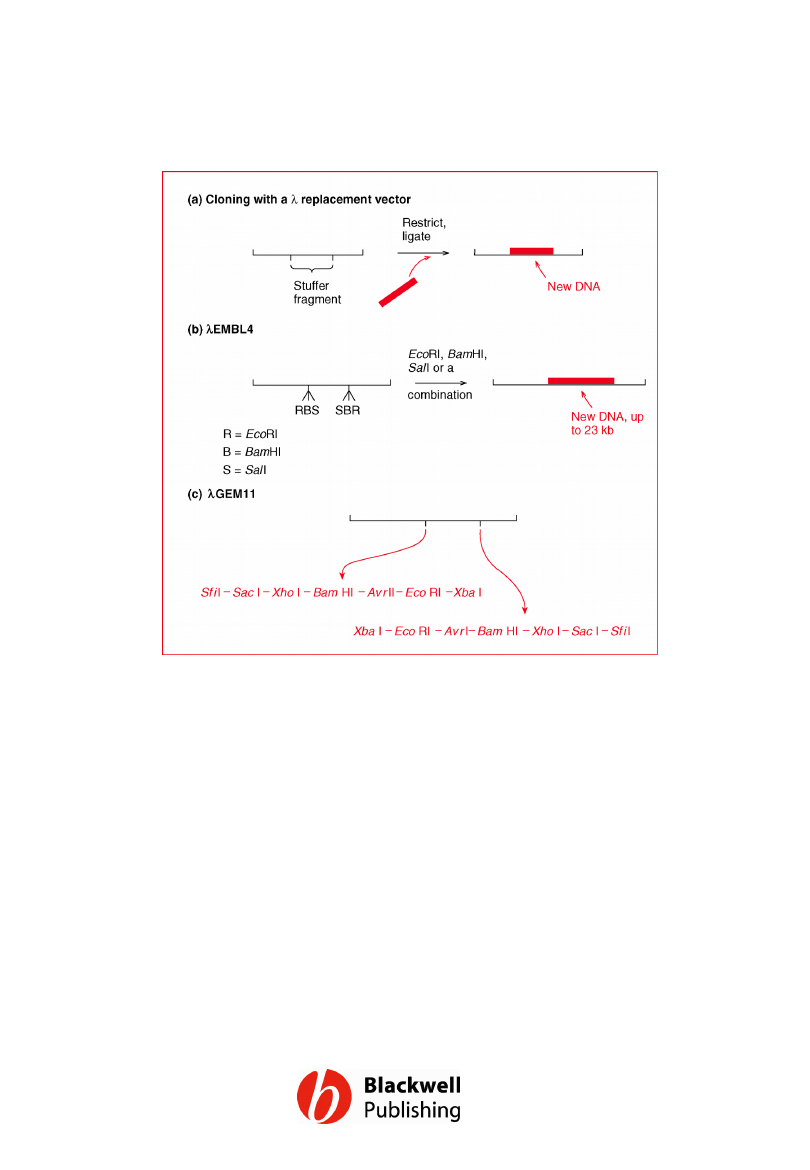
Figure 6.16 l replacement vectors. (a)
Cloning with a l replacement vector. (b)
Cloning with lEMBL4. (c) The structure of
lGEM11, showing the order of restriction sites
in the two polylinkers.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
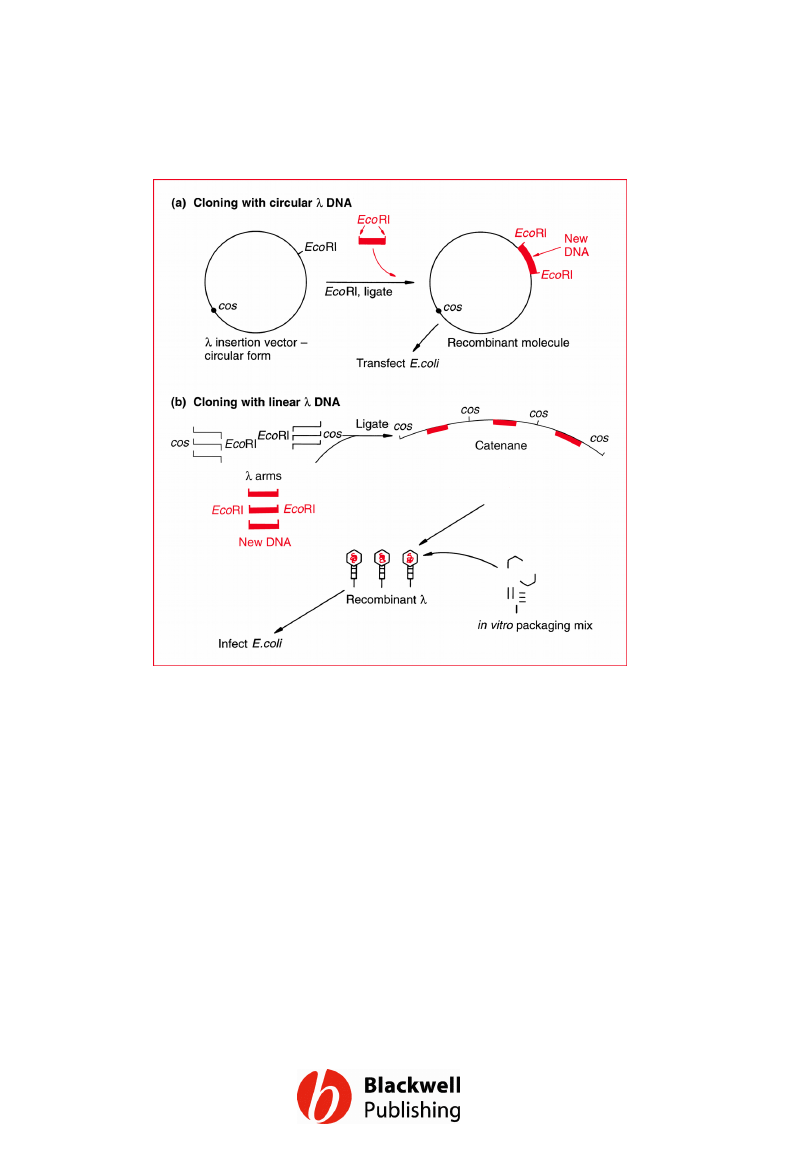
Figure 6.17 Different strategies for cloning
with a l vector. (a) Using the circular form of l
as a plasmid. (b) Using left and right arms of
the l genome, plus in vitro packaging, to
achieve a greater number of recombinant
plaques.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
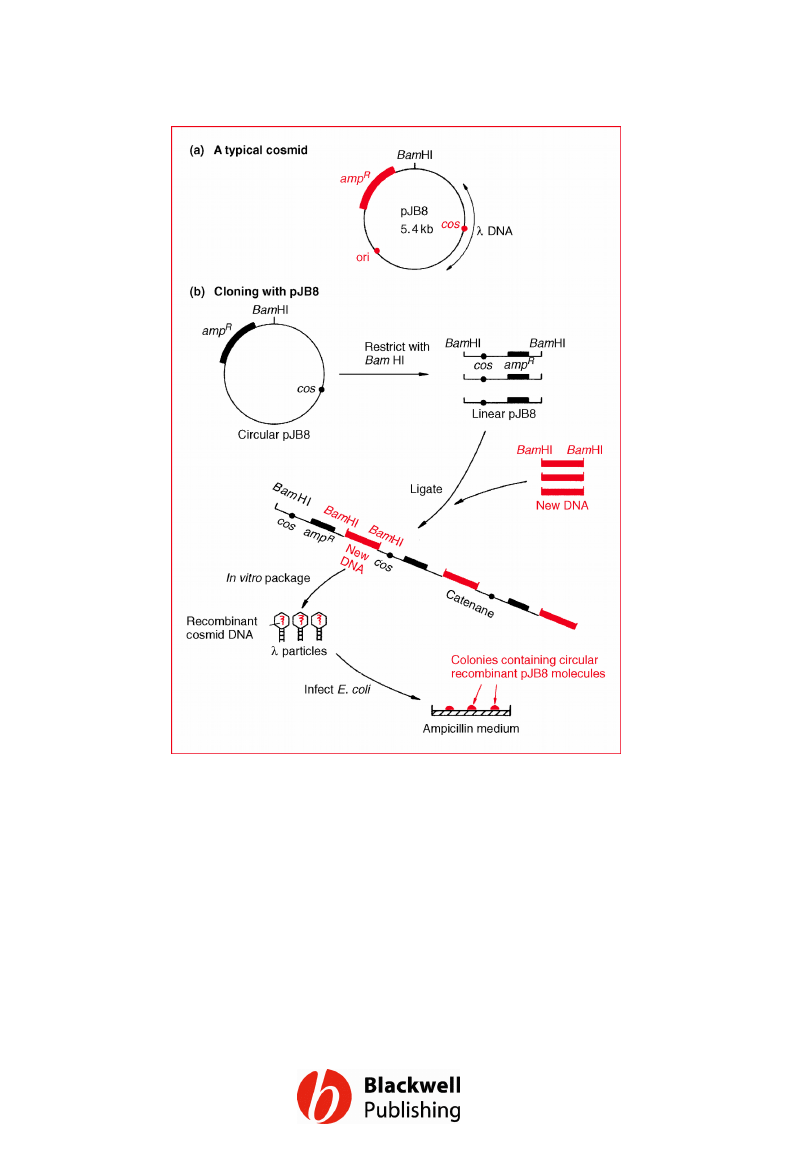
Figure 6.18 A typical cosmid and the way it
is used to clone long fragments of DNA.
Gene Cloning and DNA Analysis by T.A. Brown. © 2006 T.A.
Brown.
Document Outline
- Figure 6.1 A map of pBR322 showing the positions of the ampicillin resistance (ampR) and tetracycline resistance (tetR) genes, the origin of replication (ori) and some of the most important restriction sites.
- Figure 6.2 The pedigree of pBR322. (a) The manipulations involved in construction of pBR322. (b) A summary of the origins of pBR322.
- Figure 6.3 The pUC plasmids. (a) The structure of pUC8. (b) The restriction site cluster in the lacZ¢ gene of pUC8. (c) The restriction site cluster in pUC18. (d) Shuttling a DNA fragment from pUC8 to M13mp8.
- Figure 6.4 pGEM3Z. (a) Map of the vector. (b) In vitro RNA synthesis. R = cluster of restriction sites for EcoRI, SacI, KpnI, AvaI, SmaI, BamHI, XbaI, SalI, AccI, HincII, PstI, SphI and HindIII.
- Figure 6.5 The M13 genome, showing the positions of genes I to X.
- Figure 6.6 Construction of (a) M13mp1, and (b) M13mp2 from the wild-type M13 genome.
- Figure 6.7 Construction of M13mp7: (a) the polylinker, and (b) its insertion into the EcoRI site of M13mp2. Note that the SalI restriction sites are also recognized by AccI and HincII.
- Figure 6.8 Cloning with M13mp7 (see text for details).
- Figure 6.9 Recovery of cloned DNA from a recombinant M13mp7 molecule by restriction at the outer sites of the polylinker.
- Figure 6.10 M13mp8 and M13mp9.
- Figure 6.11 pEMBL8: a hybrid plasmid–M13 vector that can be converted into single-stranded DNA.
- Figure 6.12 The two problems that had to be solved before l cloning vectors could be developed. (a) The size limitation placed on the l genome by the need to package it into the phage head. (b) l DNA has multiple recognition sites for almost all restriction endonucleases.
- Figure 6.13 The l genetic map, showing the position of the main non-essential region that can be deleted without affecting the ability of the phage to follow the lytic infection cycle. There are other, much shorter non-essential regions in other parts of the genome.
- Figure 6.14 Using natural selection to isolate l phage lacking EcoRI restriction sites.
- Figure 6.15 l insertion vectors. P = polylinker in the lacZ¢ gene of lZAPII, containing unique restriction sites for SacI, NotI, XbaI, SpeI, EcoRI and XhoI.
- Figure 6.16 l replacement vectors. (a) Cloning with a l replacement vector. (b) Cloning with lEMBL4. (c) The structure of lGEM11, showing the order of restriction sites in the two polylinkers.
- Figure 6.17 Different strategies for cloning with a l vector. (a) Using the circular form of l as a plasmid. (b) Using left and right arms of the l genome, plus in vitro packaging, to achieve a greater number of recombinant plaques.
- Figure 6.18 A typical cosmid and the way it is used to clone long fragments of DNA.
Wyszukiwarka
Podobne podstrony:
Figures for chapter 5
Figures for chapter 12
Figures for chapter 14
Figures for chapter 10
Figures for chapter 11
Figures for chapter 8
Figures for chapter 9
Figures for chapter 2
Figures for chapter 16
Figures for chapter 13
Figures for chapter 3
Figures for chapter 7
Figures for chapter 15
Figures for chapter 1
Figures for chapter 5
Figures for chapter 12
Figures for chapter 14
Figures for chapter 10
więcej podobnych podstron