495106620

Udział
w
konferencjach naukowych: Praga 2019
HMGMT, BRCA1 AND MEG3 METHYLATION STATUS
IN TRIPLE-NEGATIVE BREAST CANCER
Sylwia Paszek1, Agnieszka Kołacińska2-3, Marcin Braun4, Ewa Kaznowska1, Dorota Jesionek-Kupnicka4, Edyta Barnaś1, Izabela Zawlik1"
^aculty of Medicine, University of Rzeszów, Rzeszów, Poland; Department of Head and Neck Cancer Surgery, Medical University of Lodź, Lodź, Poland; 3Breast Unit, Cancer Center, Copernicus Memoriał Hospital, Lodź Poland; Department of Pathology, Chair of Oncology, Medical University of Lodź, Lodź, Poland
"Corresponding author: Associate Professor Izabela Zawlik, Faculty of Medicine, University of Rzeszów, Warzywna 1A, 35-959 Rzeszów, Poland, e-mail: izazawlik@yahoo.com
BACKGROUND AND AIM: Breast cancer is one of the most common cancer in women worldwide. The most severe type of breast cancer is triple-negative breast cancer (TNBC) due to unfavorable clinical course and poor prognosis (1). The development of cancer is often associated with dysregulation of epigenetic mechanisms, including DNA methylation (2). Recent studies have shown that IncRNA (long non-coding RNA) may also affect cancerogenesis (3). One of the genes encoding IncRNA whose changes are responsible for breast cancer development is MEG3 (4). The aim of our study was to evaluate MGMT (06-methylguanine DNA methyltransferase), BRCA1 (breast cancer 1) and MEG3 fmaternally expressed 3) methylation status in TNBC.
A.
wr
_I__l__S__£__Łfi_
MUWUUWWUUM JM y UW

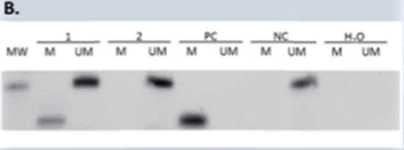
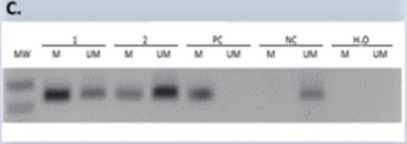
Figurę 1. Representative MSP results for the MGMT, BRCA1 and MEG3 promoter methylation. A. Presence of 80-bp PCR product indicates that the breast cancer is positive for MGMT methylation. Presence of 94-bp PCR product indicates that the breast cancer is negative for MGMT methylation. B. The product sizes of the methylated and unmethylated amplicons were 68 bp and 104 bp, respectively. C. The product sizes of the methylated and unmethylated amplicons were 227 bp. Abbreviations: M, methylated PCR product; U, unmethylated PCR product; PC, positive control - methylated control; NC, negative control -unmethylated control; MW, molecular weight marker.
EXPERIMENTAL PROCEDURĘ: In this study 44 TNBC patients were included. Genomie DNA was isolated from formalin-fixed and paraffin-embedded tissues using the QIAamp DNA FFPE Tissue Kit. The methylation status of the MGMT, BRCA1 and MEG3 promoter regions were analysed by methylation-specific PCR (MSP). We assessed the correlations between the methylation status and clinicopathological features (age, grade, tumor size, lymph nodes metastasis).
RESULTS: MGMT, BRCA1 and MEG3 promoters methylation was found in 70.4%, 61.3% and 61.3% of TNBC patients, respectively. Representative results of MSP for the promoter methylation analyzed genes are shown in Figurę 1. Moreover, we have shown that the frequency of MGMT and BRCA1 methylation is higher in older patients compared to younger patients (Table 1). Additionally, in one of TNBC patient with glandular and squamous histopathological components, it was shown that the promoters status of all analysed genes, changed from methylated to unmethylated after chemotherapy of this patient.
Table 1. Association of DNA methylation with age
|
Gene name |
Mean Age |
p - value | |
|
Unmethylated |
Methylated | ||
|
MGMT |
53.92889 |
63.54091 |
0.019438 |
|
BRCA1 |
55.00731 |
64.16015 |
0.018845 |
CONCLUSION: The high frequency of MGMT, BRCA1 and MEG3 methylation indicates that epigenetic changes are important mechanisms in breast cancer. Moreover, our results indicates that MGMT and BRCA1 methylation may have greater impact in the development of breast cancer in older patients compared to younger patients. However, the results should be confirmed in a larger number of patients.
References:
1) Siegel R, Ma J, Zou Z, Jemal A: CA: a cancer journal for dinicians 2014, Cancer Stotistics. 2014; 64:9-29. 2) Baylin SB, Jones PA. Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol. 2016; 8(9): a019505. 3) Zhou S, He Y, Yang S, et al. The regulatory roles of IncRNAs in the process of breast cancer invasion and metastasis. Biosci Rep. 2018;38(5):BSR20180772. 4) Sun LI, Li Y2, Yang B. Downregulated long non-coding RNA MEG3 in breast cancer regulates proliferation, migration and invasion by depending on p53's transcriptional activity. Biochem Biophys Res Commun. 2016;9;478(l):323-329.
FUNDING SOURCE: This study was supported 3rd World Congress on Cancer, Prague, 2019
by the University of Rzeszów.
Wyszukiwarka
Podobne podstrony:
Czynny udział»»’ konferencjach naukowych 1) 7-9.11.2000 - konferencja
ZGŁOSZENIE UDZIAŁU W KONFERENCJI NAUKOWO-SZKOLENIOWEJ(CZĘŚĆ NAUKOWA)GŁÓD I NIENASYCENIE W
illAGHDziałalność naukowa Udział w konferencjach naukowych w 2014r.: • 51
7. Udział w konferencjach naukowych 2013 A sword from Gdańsk: a technological revolution or a pagean
Zapraszamy do udziału w konferencji - 07.12.2019 r., Warszawa. Więcej
Zapraszamy do udziału w Konferencji naukowo-szkoleniowej nt.: „Rodzice pierwszymi terapeutami
XJubileuszowa Konferencja Naukowa23/11/2019 Centrum DydaktyczneUniwersytetu Medycznego ul. Pomorska
VIII. Udział w konferencjach naukowych z zaznaczeniem: funkcji organizatora, współorganizatora, form
Ą Konferencja Naukowo-WarsztatowaHortiterapia jako element wspomagającyleczenie tradycyjne 20 sierpn
IV Konferencja Naukowo-Techniczna, Kołobrzeg, 28-29 maja 2019 r.POMIARY I DIAGNOSTYKA W SIECIACH
• Odbyło słę 11 konferencji naukowych, w których udział wzięło 1118 osób, w tym 51
PEHIN6 ZY1RO SUWAKI A XXVIII KONFERENCJA NAUKOWO-TECHNICZNA 22.-24.5.2019 w
konferencjach naukowych Warszawa 2019Influence of polymorphisms in APOE, APOC1, and ACE ge
OGÓLNOPOLSKA KOGN1TYWISTYC7.NA KONFERENCJA NAUKOWACEREBRUMCOGNITA l Lublin, 17 maja 2019 r.
więcej podobnych podstron