
Journal of Biotechnology 92 (2001) 27 – 35
Multidrug resistance as a dominant molecular marker in
transformation of wine yeast
Z. Kozovska
a
, A. Maraz
b
, I. Magyar
c
, J. Subik
a,
*
a
Department of Microbiology and Virology, Faculty of Natural Sciences, Comenius Uni
6ersity, Mlynska dolina B-
2
,
84215
Bratisla
6a, Slo6akia
b
Department of Microbiology and Biotechnology, Faculty of Food Sciences, Szent Ist
6an Uni6ersity, Somloi ut
14
-
16
,
1118
Budapest, Hungary
c
Department of Enology, Faculty of Food Sciences, Szent Ist
6an Uni6ersity, Somloi ut
14
-
16
,
1118
Budapest, Hungary
Received 2 February 2001; received in revised form 7 June 2001; accepted 20 June 2001
Abstract
Pure wine yeast cultures are increasingly used in winemaking to perform controlled fermentations and produce wine
of reproducible quality. For the genetic manipulation of natural wine yeast strains dominant selective markers are
obviously useful. Here we demonstrate the successful use of the mutated PDR
3 gene as a dominant molecular marker
for the selection of transformants of prototrophic wine yeast Saccharomyces cere
6isiae. The selected transformants
displayed a multidrug resistance phenotype that was resistant to strobilurin derivatives and azoles used to control
pathogenic fungi in agriculture and medicine, respectively. Random amplification of DNA sequences and elec-
trophoretic karyotyping of the host and transformed strains after microvinification experiments resulted in the same
gel electrophoresis patterns. The chemical and sensory analysis of experimental wines proved that the used
transformants preserved all their useful winemaking properties indicating that the pdr
3-9 allele does not deteriorate
the technological properties of the transformed wine yeast strain. © 2001 Elsevier Science B.V. All rights reserved.
Keywords
:
Dominant marker; PDR
3
gene; Multidrug resistance; Transformation; Wine yeast
www.elsevier.com/locate/jbiotec
1. Introduction
Saccharomyces cere
6isiae strains belong to the
most widely used yeast in winemaking. They rep-
resent a dominant part of the microflora of grape
juices and young wines. Nowadays, many wine-
makers use the pure wine yeast cultures to per-
form controlled must fermentation resulting in
production of wines of reproducible quality
(Snow, 1983; Malik et al., 1990; Nemecek et al.,
1990; Querol et al., 1992). The yeast is selected for
desirable characteristics like ethanol and osmotol-
erance, high fermentation activity or cold and
chemoresistance (Snow, 1983; Moreno et al.,
1991). The improvement of the traits that play a
role in must fermentation is one of the most
important biotechnological challenges in wine
yeast.
* Corresponding author. Tel.: + 42-12-6029-6631; fax: +
42-12-6542-9064.
E-mail address
:
subik@fns.uniba.sk (J. Subik).
0168-1656/01/$ - see front matter © 2001 Elsevier Science B.V. All rights reserved.
PII: S 0 1 6 8 - 1 6 5 6 ( 0 1 ) 0 0 3 4 6 - 7

Z. Kozo
6ska et al.
/
Journal of Biotechnology
92 (2001) 27 – 35
28
The advent of molecular techniques and their
applications to the study of wine fermentation
provide a useful tool for genetic improvement of
wine yeast strains. However, they are pro-
totrophic and homothallic with a higher degree of
ploidy. Therefore, dominant selectable markers
are required for a successful transformation of
such strains. Several dominant drug resistance
markers have been developed for use in S. cere-
6isiae (reviewed in Van den Berg and Steensma,
1997; Lackova and Subik, 1998) from which the
G418 resistance (Wach et al., 1994) have achieved
a wide-spread use.
Recent studies carried out in our laboratory
showed that both multicopy and centromeric vec-
tors bearing the mutant pdr
3
-
9
allele provide a
tool for the direct selection of transformants of
laboratory and polyploid industrial strains of bak-
er’s yeast (Nourani et al., 1997; Lackova and
Subik, 1999). The Pdr3p mutant protein acts as a
gain-of-function transcriptional activator respon-
sible for the establishment of multidrug resistance
phenotype due to the overexpression of several
membrane drug efflux pumps (Nourani et al.,
1997; Michalkova-Papajova et al., 2000; DeRisi et
al., 2000).
In this work, the pdr
3
-
9
gene was used as the
dominant selectable marker for the transforma-
tion of a natural wine strain of S. cere
6isiae.
2. Materials and methods
2
.
1
. Strains and culture media
The ethanol tolerant S. cere
6isiae wine yeast
strains of the FV series-originated from the vine-
growing regions in Slovakia (Malik et al., 1996)
were used for transformation and microvinifica-
tion experiments. Escherichia coli strain XL1 Blue
was used for plasmid amplification. Yeast cells
were grown at 30 °C in complex glucose (YPD)
and complex glycerol (YEG) medium (1% yeast
extract, 2% bacteriological peptone, 2% glucose or
2% glycerol) or in minimal (YNB) medium (0.67%
yeast nitrogen base without amino acids, 2% glu-
cose or 2% glycerol plus 1% ethanol). E. coli cells
were grown at 37 °C in the LB medium (1%
tryptone, 1% NaCl, 0.5% yeast extract, pH 7.5).
The plasmid propagation medium was supple-
mented with 100
mg ml
− 1
of ampicillin. The me-
dia were solidified with 2% bactoagar.
2
.
2
. Plasmid and transformation protocols
The mutant pdr
3
-
9
allele under the control of
its promoter was cloned on centromeric plasmid
pFL38/pdr3-9 (ARS
1
CEN
4
URA
3
ori amp
R
)
(Lackova and Subik, 1999). E. coli were trans-
formed and all DNA manipulations were carried
out as described in Sambrook et al. (1989). The
yeast strain S. cere
6isiae FV1 was transformed by
electroporation (Thompson et al., 1998) and the
resulting transformants were selected for drug re-
sistance. Plasmid DNA was extracted from yeast
cells as described in Alister and Ward (1990).
2
.
3
. Drug sensiti
6ity assays
The sensitivity of yeast cells to drugs was as-
sayed by determination of growth inhibition zones
(Michalkova-Papajova et al., 2000) using antifun-
gal susceptibility discs (ITEST plus Ltd, Czech
Republic) and by measuring the minimal in-
hibitory concentrations (MIC) on the solid
medium containing different drug concentrations
(Nourani et al., 1997). In the case of mitochon-
drial
inhibitors
(chloramphenicol,
mucidin,
azoxystrobin and kresoxim-methyl) the media
contained 2% glycerol and 1% ethanol instead of
glucose. Drug resistance was scored after 6 days
of growth at 30 °C.
2
.
4
. Determination of plasmid loss
Transformants were grown in liquid YPD
medium and re-inoculated to fresh medium to
initial concentration of 1 × 10
5
cell ml
− 1
each 24
h. Appropriate dilutions of the cell suspension
were plated onto solid YPD medium. After 2 days
of growth at 30 °C, colonies were replica-plated
onto YPD medium containing cycloheximide (1
mg ml
− 1
) and two YPG media containing chlo-
ramphenicol (3 mg ml
− 1
) and mucidin (0.2
mg
ml
− 1
), respectively, for the evaluation of the plas-
mid-containing fraction.

Z. Kozo
6ska et al.
/
Journal of Biotechnology
92 (2001) 27 – 35
29
2
.
5
. RAPD-PCR analysis
2
.
5
.
1
. DNA isolation
About 1.5 ml of the overnight shaken culture in
YPD medium was centrifuged and the cells were
re-suspended in 200
ml breaking buffer (2% Tri-
ton-X 100, 1% SDS, 100 mM NaCl, 10 mM
Tris – HCl, 1 mM EDTA, pH 8). After addition of
0.3 g glass beads (0.5 mm diameter) and 200
ml
PCIA
(phenol:chloroform:isoamylalcohol =
25:24:1) the mixture was vortexed for 3 min. 200
ml TE buffer (10 mM Tris–HCl, 1 mM EDTA,
pH 8) was added by gently mixing and the tube
was centrifuged for 5 min. DNA in supernatant
was precipitated with ethanol and treated by
RNase (10
mg ml
− 1
) in 100
ml TE buffer.
2
.
5
.
2
. PCR amplification by random primers
The PCR cocktail (30
ml) contained 50 pM
primer (P1: 5
%-GTGGTGGTG-3%, P2: 5%-CGCGT-
GCCCA-3
%, OPE7: 5%-AGATGCAGCC-3%), 25
mM MgCl
2
, 20 mM dNTPs, 1
mg genomic DNA
and 1.5 U Taq polymerase. Amplification was
performed as follows, 2 min at 95 °C; 35 cycles at
95 °C for 30 s; at 40 °C (P1) or 38 °C (P2 and
OPE7) for 30 s and 72 °C for 3 min. Final
extension, 72 °C for 7 min.
2
.
6
. Electrophoretic karyotyping
Intact chromosomes were prepared by lysis of
protoplasts embedded into low melting point
agarose according to the method of Carle and
Olson (1985). Separation of chromosomes was
performed by Rotaphor Type IV (Biometra) in
0.9% agarose according to the instruction of the
producer.
2
.
7
. Micro
6inification experiments
White must of Furmint grape from Tokaj re-
gion, with 258 g l
− 1
reducing sugar, 9.2 g l
− 1
titratable acid, 4/62 g l
− 1
free/total sulphur diox-
ide, pH 3.04 was sterilised by filtration and inocu-
lated with the yeast strains at a concentration of
10
6
cells/ml. Fermentation was performed in 600
ml must volume (in 700 ml bottle), three parallels
at 18 °C. Progress of fermentation was monitored
by measuring the refraction of the must (Abbe-
type Refractometer, Zeiss). At the end of the
fermentation, wines were centrifuged at 6000 rpm
for 10 min to remove yeast cells and kept at 4 °C
prior analysis. Enological parameters of the wines
were determined as described in Querol et al.
(1990). Sensory analyses were done by profes-
sional judges.
3. Results
3
.
1
. Transformation for drug resistance of natural
wine yeast
The wine yeast strain S. cere
6isiae FV1 used in
the transformation experiments was isolated from
Mu¨ller – Thurgau wine originating from the vine-
growing region Skalica – Zahorie. It was selected
from the five strains of the FV series (Malik et al.,
1996) due to its highest susceptibility to mucidin
and chloramphenicol, the antibiotics inhibiting
the respiration and protein synthesis in mitochon-
dria, respectively, and also to cycloheximide,
which is a potent inhibitor of cytoplasmic protein
synthesis.
After subcloning, the prototrophic strain FV1
was transformed by electroporation with plasmid
pFL38/pdr3-9 carrying the gain-of-function allele
of the PDR
3
gene as the dominant selectable
marker (Lackova and Subik, 1999). Transfor-
mants were selected for resistance to either cyclo-
heximide (1
mg ml
− 1
) or chloramphenicol (3 mg
ml
− 1
). In the transformed strain the frequency of
resistance of spontaneous mutants to chloram-
phenicol or cycloheximide was at least six times
lower (Table 1). The range of transformation
efficiencies in repeated experiments was between 1
and 4 × 10
3
per
mg DNA. Transformants were
distinguished from the spontaneous mutants on
their ability to display multidrug resistance and
provide 9.3 kb plasmid DNA conferring ampi-
cillin resistance in E. coli and giving the expected
DNA fragments after restriction with HindIII or
HpaI (Nourani et al., 1997). In isolated transfor-
mants the plasmid conferred a high level of resis-
tance to different drugs (Table 1) corroborating
the results obtained by industrial strains of bak-
er’s yeast.
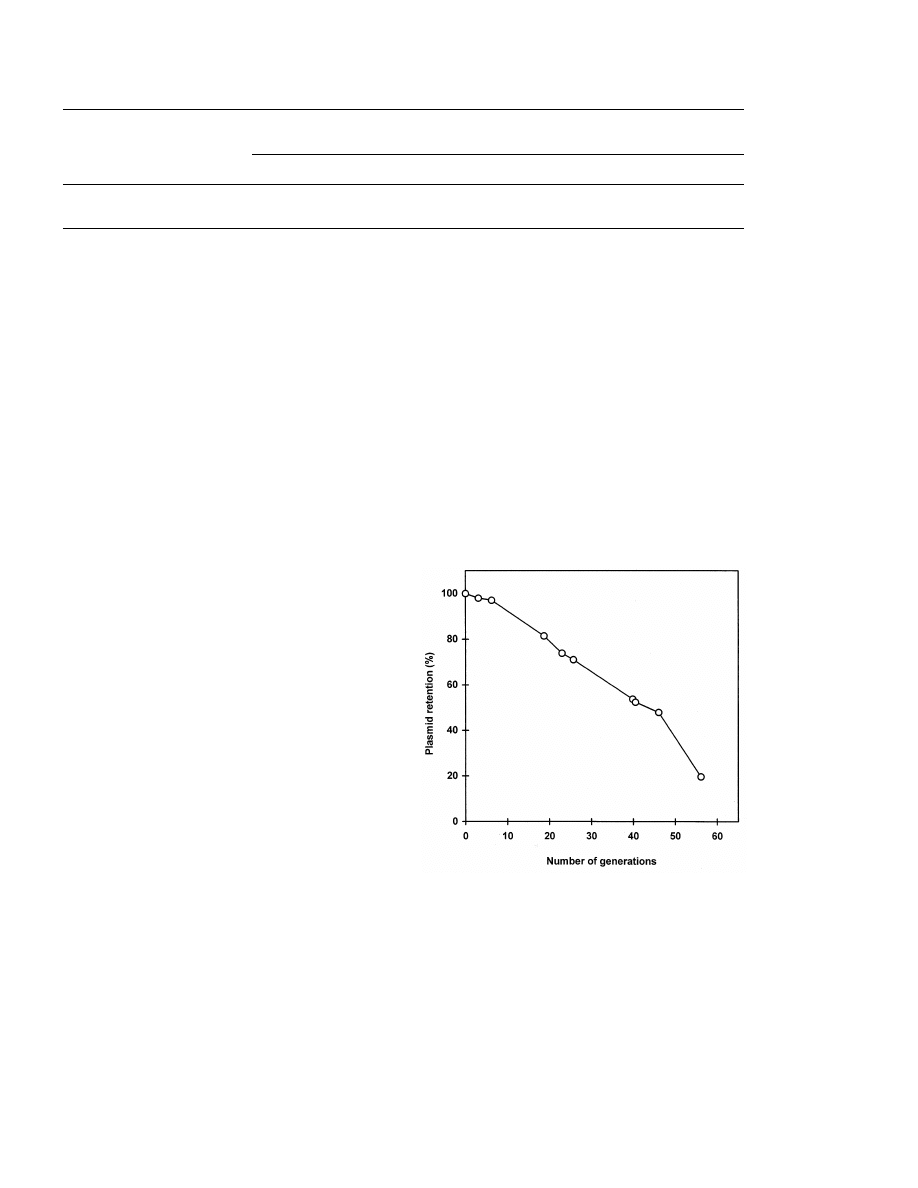
Z. Kozo
6ska et al.
/
Journal of Biotechnology
92 (2001) 27 – 35
30
Table 1
Transformation of natural wine yeast strain with a centromeric plasmid carrying the pdr
3
-
9
mutant allele
S. cere
6isiae
Plasmid
Frequency of spontaneous mutants Minimal inhibitory concentration
and transformants selected as
Cyh
R
Cmp
R
CYH (
mg ml
−1
)
CMP (mg ml
−1
)
MUC (
mg ml
−1
)
3.2×10
−7
2.5×10
−6
FV1
0.6
No
2
0.1
pFL38/pdr3-9
FV1
1.1×10
−5
1.5×10
−5
2.5
4
0.6
Abbreviations: Cyh
R
, cycloheximide resistant; Cmp
R
, chloramphenicol resistant; CYH, cycloheximide; CMP, chloramphenicol;
MUC, mucidin.
In spite of the present centromere, moderate
loss of plasmid from transformants was observed
during mitotic growth in the absence of any drug
in complex medium containing glucose (Fig. 1).
However, 50% of yeast population still showed
multidrug resistance (Cyh
R
Cmp
R
Muc
R
) even
after 40 cell generations indicating a sufficiently
high stability of plasmid in transformants of the
natural wine yeast strain.
3
.
2
. Drug resistance pattern, experimental wine
production and molecular fingerprinting
The PDR
3
gene is known to regulate subsets of
genes involved in multidrug resistance and other
biological processes (DeRisi et al., 2000). There-
fore, we determined the susceptibility of the natu-
ral wine yeast strain S. cere
6isiae FV1 and its
transformants to several antifungal agents used in
agriculture and medicine, and assessed the enolog-
ical parameters of the wines obtained by both
yeast strains.
As is shown in Table 2, transformants carrying
the pdr
3
-
9
allele exhibited differential degrees of
susceptibility to fungicides used to control the
filamentous fungi in the vineyards. Clear resis-
tance was observed to the derivatives of strobil-
urin (Mucidin, Discus, Quadris) and certain
azoles (Hattrick, Topas). On the other hand,
transformants were found to be more sensitive to
Anvil 5SC and Vectra 10SC, or exhibited no
changes to other commercial fungicides tested. An
increased level of resistance of transformants was
also observed for several antimycotics, that in the
case of imidazoles was more pronounced on com-
plex than on minimal medium (Table 3).
To show how the natural vinification ability of
the FV1 strain is conserved in its transformants
bearing the pdr
3
-
9
allele we carried out mi-
crovinification experiments with two yeast strains.
The analytic parameters of the wines obtained
and their sensory analysis are shown in Table 4
and Table 5, respectively. The results from three
experiments clearly show that no significant dif-
ferences were observed between the natural yeast
strain and its pdr
3
-
9
transformants.
Possible changes in the length of chromosomes
as the consequence of the interaction of recombi-
nant plasmids and chromosomes during vinifica-
Fig. 1. Stability of plasmid pFL38/pdr3-9 in the population of
the wine yeast transformants growing in complex medium with
glucose under non-selective conditions.

Z. Kozo
6ska et al.
/
Journal of Biotechnology
92 (2001) 27 – 35
31
Table 2
Susceptibility to different fungicides of the S. cere
6isiae strain FV1 and its transformants TFV1
Fungicide
Commercial name
Minimal inhibitory concentration (
mg ml
−1
)
FV1
Active compounds
TFV1
Kresoxim-methyl
Discus
0.05
0.6
0.04
Quadris
0.4
Azoxystrobin
0.10
0.6
Strobilurin A
Mucidin
400
Anvil 5SC
200
Hexaconazole
Bumper 25C
Propiconazole
200
200
50
75
Hattrick
Tebuconazole plus tolyfluanide
200
Penconazole
600
Topas
Bromuconazole
Vectra 10SC
75
50
100
Acrobat
100
Dimethomorh plus mancozeb
8000
Fosethyl-Al
8000
Aliette
Folpet
Folpan 80WG
50
50
50
50
Novozir MN80
Mancozeb
100
Metiram
100
Polyram WG
Metalaxyl plus mercury oxychloride
Ridomyl Plus 48WP
5000
5000
\16 000
\16 000
Sumilex 50WP
Procymidone
16 000
16 000
Sulphur
Thiovit
Table 3
Susceptibility to antimycotics of the S. cere
6isiae strain FV1 and its transformant TFV1 determined on complex and minimal
medium containing glucose
Medium
Saccharomyces
Diameter of inhibition zone (mm)
cere
6isiae strain
Azoles
Polyenes
ECO
MCZ
CLO
KET
BIF
FLU
ITR
PIM
AMB
NYS
5-FC
27
19
18
13
10
YPD
0
FV1
0
20
0
19
0
15
12
0
0
0
0
0
18
0
19
0
TFV1
28
28
20
8
17
0
FV1
11
YNB
22
12
27
30
26
25
10
0
11
0
0
24
10
25
40
TFV1
Abbreviations: ECO, econazole; MCZ, miconazole; CLO, clotrimazole; KET, ketoconazole; BIF, bifonazole; FLU, fluconazole;
ITR, itraconazole; PIM, pimaricin; AMB, amphotericin B; NYS, nystatin; 5-FC, 5-fluorocytosin.
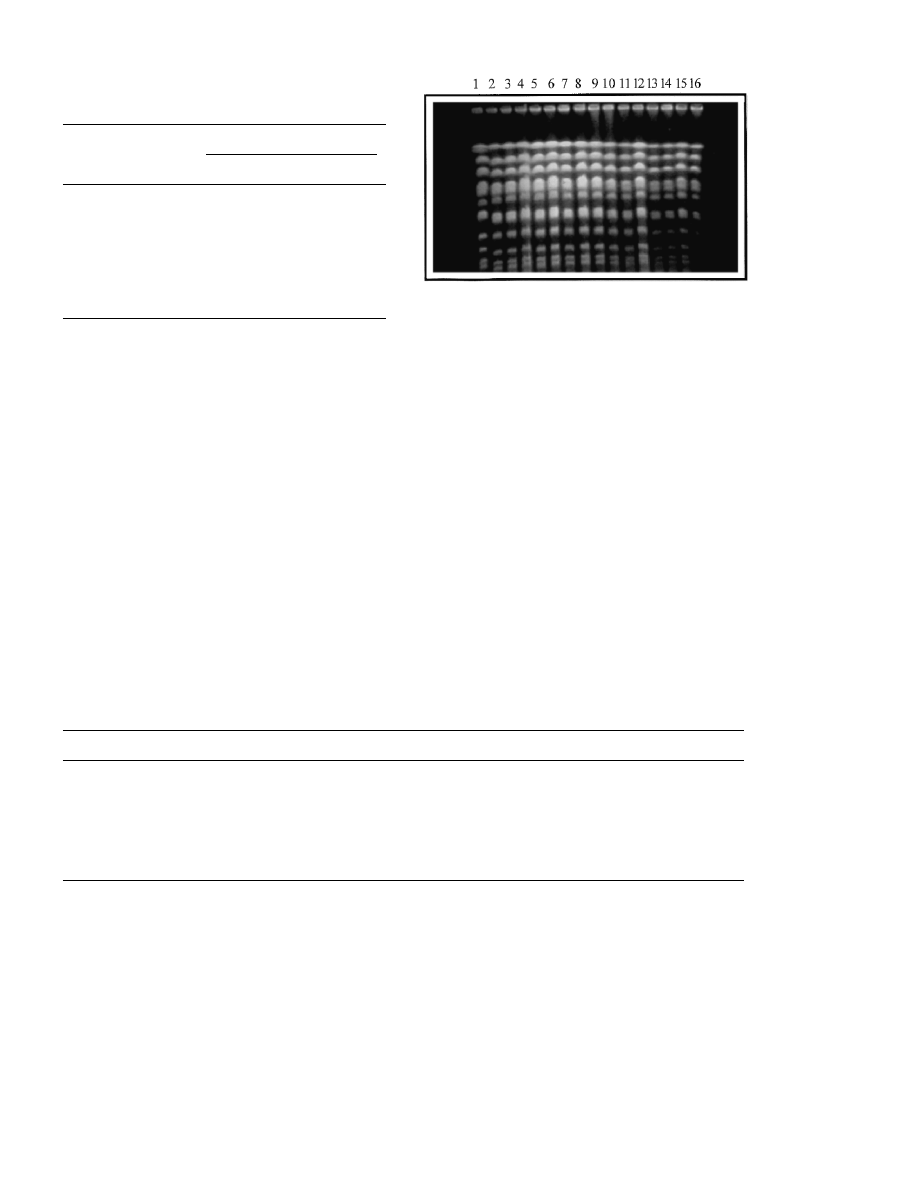
tion was checked by electrophoretic separation of
the intact chromosomes. Results shown in Fig. 2
indicate that no detectable changes occurred dur-
ing propagation of the cells, either they lost the
cycloheximid resistance (TFV1/S isolates) or they
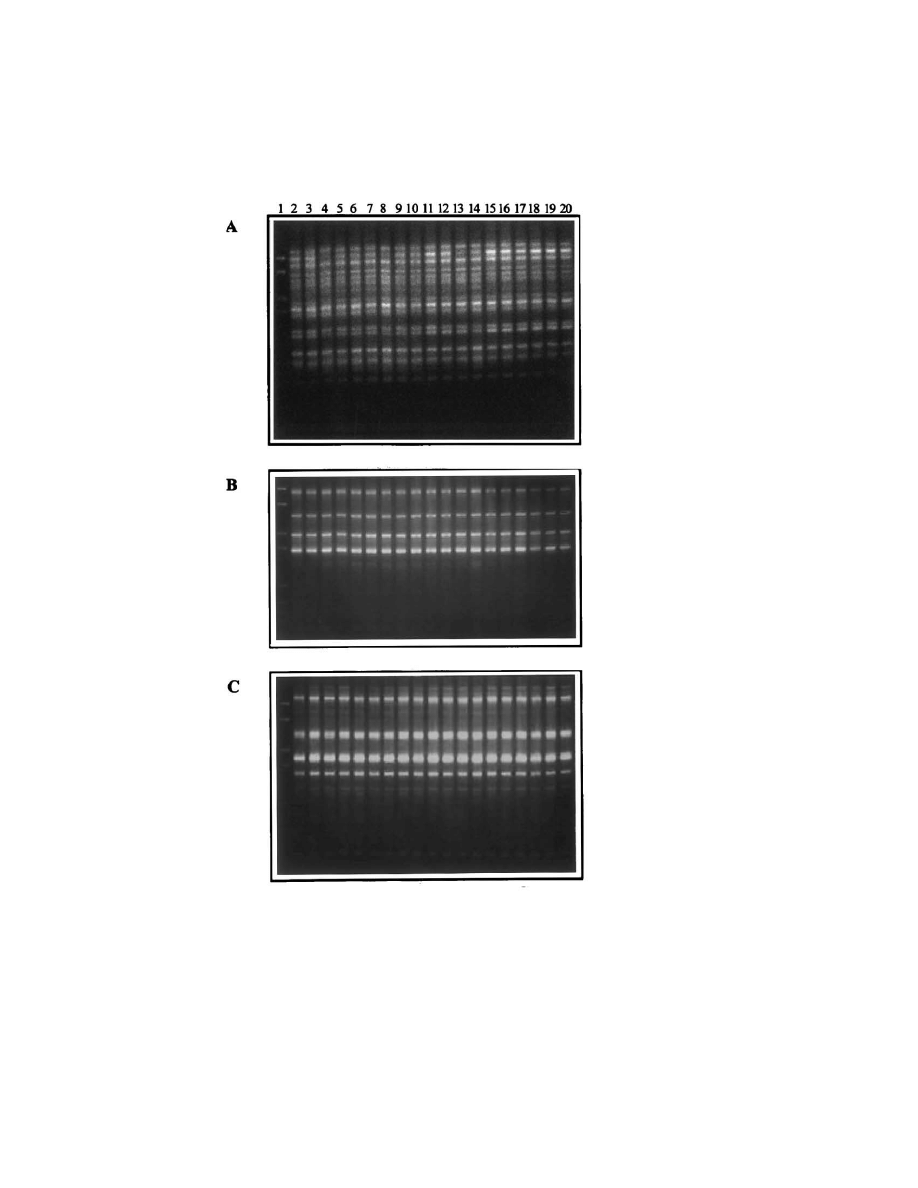
retained it (TFV1/R isolates). Random amplifica-
tion of the genomic DNA sequences by three
different primers also showed that no alteration in
the length or number of the amplified fragments
took place (Fig. 3).
The data obtained here indicate that multiple
drug resistance conferred by mutant pdr
3
-
9
allele
is a valuable dominant selectable marker that can
be successfully used to transform natural S. cere-
6isiae wine strains without deterioration of their
enological properties.
Z. Kozo
6ska et al.
/
Journal of Biotechnology
92 (2001) 27 – 35
32
Table 4
Chemical analysis of experimental wines after completion of
alcoholic fermentation
Enological parameter
Value
FV1
TFV1
1.67
90.13
4.63
93.71
Reducing sugar (g l
−1
)
Ethanol (%, v/v)
15.05
90.13
14.92
90.11
49.7
918.67
61.3
93.97
Acetaldehyde (mg l
−1
)
29.70
90.34
Total extract (g l
−1
)
32.80
94.33
8.27
90.13
8.70
90.11
Total acidity (g l
−1
)
Volatile acidity (g l
−1
)
0.62
90.06
0.67
90.09
3.17
90.01
3.14
90.01
pH
Colour (A
420
)
0.17
90.01
0.12
90.02
The main values of triplicates and their confidence intervals at
P = 0.05.
Fig. 2. Electrophoretic karyograms of the host (FV1) and the
transformant (TFV1) strains isolated at the end of the mi-
crovinification. Lane 1, marker strain (S288c); lane 2, FV1;
lanes 3 – 8, TFV1/S1 – TFV1/S6; lane 9, TFV1; lanes 10 – 16,
TFV1/R1 – TFV1/R7.
4. Discussion
In this study we successfully used the pdr
3
-
9
mutant allele on the centromeric plasmid for di-
rect selection of transformants of the natural wine
yeast S. cere
6isiae strain FV1 for resistance to
chloramphenicol and cycloheximide, respectively.
Equally effective as pdr
3
-
9
were the other gain-of-
function pdr
3
alleles, like the pdr
3
-
7
(Nourani et
al., 1997) and pdr
3
-
20
(Simonics et al., 2000)
(data not shown). The pdr
3
-
9
dominant selectable
marker encodes the mutant transcription factor
Pdr3p, with Y276H amino acid substitution,
which is responsible for an increased expression of
several drug efflux pumps (Nourani et al., 1997;
DeRisi et al., 2000). The substrate specificity of
these membrane transporters is relatively broad
(Kolaczkowski et al., 1998; Bauer et al., 2000).
Therefore, the prototrophic wine yeast transfor-
mants were expected to possess an increased level
of resistance to many structurally and functionally
unrelated drugs. In fact, resistance of transfor-
mants to several antifungal agents used against
phytopathogenic fungi and human fungal patho-
gens was clearly demonstrated (Tables 2 and 3)
even in the presence of at least two copies of the
chromosomal wild-type PDR
3
gene in the strain
FV1 (Malik et al., 1996). Therefore, after the
efficient transformation using the improved proto-
col for the preparation of yeast cells for electropo-
ration (Thompson et al., 1998), transformants
Table 5
Sensory analysis of experimental wines produced by the S. cere
6isiae strain FV1 and its transformants TFV1
Flavour/taste
Odour
Total scores
Colour
Wines
3.0
5.0
FV1
3.4
11.4
11.7
4.9
3.1
3.5
4.7
3.2
3.2
11.1
TFV1
4.9
3.3
3.6
11.7
4.9
3.3
11.8
3.6
3.0
3.4
3.8
12.1
0.7136
0.7800
0.4046
Level of significance (P)
0.6765
The six wine samples (2 strains×3 parallels) were examined by five judges in one trial. The identity of the wines examined in
randomised order was not known by the judges. Data were evaluated by one-way analysis of variance where the individual scores
of the judges were considered ‘repetition’.
Z. Kozo
6ska et al.
/
Journal of Biotechnology
92 (2001) 27 – 35
33
may be directly selected for resistance to any drug
known to be actively pumped out of the cells
carrying a gain-of-function mutation in either
PDR
1
or PDR
3
gene.
Since the prototrophic strain FV1 sporulate
with the frequency of about 50% (Malik et al.,
1996), at least half of the resulting spores are
expected to harbour the centromeric plasmid and
Fig. 3. RAPD-PCR patterns of the transformants isolated at the end of the microvinification. A-primer P1, B-primer P2, C-primer
OPE7. Lane 1, DNA size marker; lane 2, TFV1; lanes 3 – 14, TFV1/R1 – TFV1/R12; lanes 15 – 20, TFV1/S1 – TFV1/S6.

Z. Kozo
6ska et al.
/
Journal of Biotechnology
92 (2001) 27 – 35
34
express multidrug resistance phenotype. This cre-
ates an opportunity for wine yeast strain im-
provement by genetic hybridisation using the
mitochondrial (rho
−
) respiratory deficient pdr
3
-
9
spores (induced by ethidium bromide) and res-
piratory competent (rho
+
) drug sensitive partner
cells. After mass mating of spore suspensions the
resulting hybrids can be selected as a respiratory
competent and drug resistant cell population that
will be able to sporulate.
The PDR
3
gene controls (directly or indirectly)
the transcription of numerous genes involved not
only in multidrug resistance phenomenon but
also in sugar transport, organelle function, phos-
pholipid metabolism, cell wall biosynthesis and
resistance to physical stresses (DeRisi et al.,
2000). The changes in the expression of such a
large set of genes could also modify some tech-
nological properties of the transformed yeast
strains. However, as shown in Tables 4 and 5,
transformants of wine yeast carrying centromeric
plasmids with pdr
3
-
9
mutant allele produced ex-
perimental wines of the same quality as their
parental natural strain FV1. This opens the way
to construct recombinant wine yeast strains con-
taining various enologically interesting enzymes
(Pretorius, 2000). Nevertheless, single gain-of-
function mutant alleles of the PDR
3
gene may
also have a beneficial effects for transformants of
industrial strains. This has already been shown
for the mutant alleles of homologous PDR
1
gene
in clotrimazole-resistant sake yeast that showed
improved fermentation activity in sake mash
even in the final fermentation stage (Mizoguchi
et al., 1999).
Acknowledgements
We thank Professor F. Malik for the wine
yeast strains, Dr J. Markovic for the samples of
commercial fungicides and Dr D. Hanson for
critical reading of the manuscript. This work was
supported by grants from the Slovak Grant
Agency of Science (VEGA), Slovak Ministry of
Education and Howard Hughes Medical Institute
(USA).
References
Alister, C., Ward, L., 1990. Single step purification of shuttle
vectors from yeast for high frequency back-transformation
into E. coli. Nucl. Acid Res. 18, 5318 – 5324.
Bauer, B.E., Wolfger, H., Kuchler, K., 2000. Inventory and
function of yeast ABC proteins: sex, stress, pleiotropic
drug and heavy metal resistance. Biochim. Biophys. Acta
1461, 217 – 236.
Carle, G.F., Olson, M.V., 1985. An electrophoretic karyotype
for yeast. Proc. Natl. Acad. Sci. USA 82, 3756 – 3760.
DeRisi, J., Van den Hazel, B., Marc, P., Balzi, E., Brown, P.,
Jacq, C., Goffeau, A., 2000. Genome microarray analysis
of transcriptional activation in multidrug resistance yeast
mutants. FEBS Lett. 470, 156 – 160.
Kolaczkowski, M., Kolaczkowska, A., Luczynski, J., Witek,
S., Goffeau, A., 1998. In vivo characterisation of the drug
resistance profile of the major ABC transporters and other
components of the yeast pleiotropic drug resistance net-
work. Microb. Drug Resist. 4, 143 – 158.
Lackova, D., Subik, J., 1998. Dominant selectable vector
markers in transformation of the yeast Saccharomyces
cere
6isiae (In Slovak). Biol. Listy 63, 259–275.
Lackova, D., Subik, J., 1999. Use of mutated PDR
3
gene as a
dominant selectable marker in transformation of pro-
totrophic yeast strains. Folia Microbiol. 44, 171 – 176.
Malik, F., Kukanova, A., Buchtova, V., Krasny, S., Krapek,
J., 1990. Biotechnological properties of yeast isolated for
secondary wine fermentation use-5th part: Application of
isolated yeast in production units (in Slovak). Kvas. Prum.
36, 36 – 38.
Malik, F., Sitorova, S., Vollek, V., Linczenyiova, K., 1996.
Microbiological and cultivation properties of wine yeast
Saccharomyces cere
6isiae of the FV series. Biolo´gia 51,
629 – 634.
Michalkova-Papajova, D., Obernauerova, M., Subik, J., 2000.
Role of the PDR gene network in yeast susceptibility to the
antifungal
antibiotic
mucidin.
Antimicrob.
Agents
Chemother. 44, 418 – 420.
Mizoguchi, H., Watanabe, M., Nishimura, A., 1999. Charac-
terisation of a PDR1 mutant allele from a clotrimazole-re-
sistant sake yeast mutant with improved fermentative
activity. J. Biosci. Bioeng. 88, 20 – 25.
Moreno, J.J., Millan, C., Ortega, J.M., Medina, M., 1991.
Analytical differentiation of wine fermentations using pure
and mixed yeast cultures. J. Ind. Microb. 7, 181 – 190.
Nemecek, F., Strakova, K., Krasny, S., 1990. Utilisation of
active dry wine yeast in sparkling wine technology. (In
Slovak). Kvas. Prum. 36, 70 – 72.
Nourani, A., Papajova, D., Delahodde, A., Jacq, C., Subik, J.,
1997. Clustered amino acid substitutions in the yeast tran-
scription regulator Pdr3p increase pleiotropic drug resis-
tance and identify a new central regulatory domain. Mol.
Gen. Genet. 256, 397 – 405.
Pretorius, I.S., 2000. Tailoring wine yeast for the new millen-
nium: novel approaches to the ancient art of winemaking.
Yeast 16, 675 – 729.

Z. Kozo
6ska et al.
/
Journal of Biotechnology
92 (2001) 27 – 35
35
Querol, A., Jime´nez, M., Huerta, T., 1990. Microbiological
and enological parameters during fermentation of musts
from poor and normal grape-harvest in the region of
Alicante (Spain). J. Food Sci. 55, 114 – 122.
Querol, A., Barrio, E., Huerta, T., Ramo´n, D., 1992. Strain
for use as dry yeast in Fermentation of Alicante wines:
Selection and DNA pattern. J. Food Sci. 57, 183 – 185.
Sambrook, J., Fritsch, E.F., Maniatis, T., 1989. Molecular
Cloning: a Laboratory Manual. Harbor Laboratory Press,
Cold Spring Cold Spring Harbour, NY.
Simonics, T., Kozovska, Z., Michalkova-Papajova, D., Dela-
hodde, A., Jacq, C., Subik, J., 2000. Isolation and molecu-
lar characterization of the carboxy-terminal pdr
3
mutants
in Saccharomyces cere
6isiae. Curr. Genet. 38, 248–255.
Snow, R., 1983. Genetic manipulation of wine yeast. In:
Spencer, J.F.T., Spencer, D.M., Smith, A.R.W. (Eds.),
Yeast
Genetics
Fundamental
and
Applied
Aspects.
Springer, Berlin, p. 439.
Thompson, J.R., Register, E., Curotto, J., Kurtz, M., Kelly,
R., 1998. An improved protocol for the preparation of
yeast cells for transformation by electroporation. Yeast 14,
565 – 571.
Van den Berg, M.A., Steensma, H.Y., 1997. Expression cas-
settes of formaldehyde and fluoroacetate resistance, two
dominant markers in Saccharomyces cere
6isiae. Yeast 13,
551 – 559.
Wach, A., Brachat, A., Pohlmann, R., Philippsen, P., 1994.
New heterologous modules for classical or PCR-based
gene disruptions in Saccharomyces cere
6isiae. Yeast 10,
1793 – 1808.
Wyszukiwarka
Podobne podstrony:
Screening for effectors that modify multidrug resistance in yeast
Pdr1 regulates multidrug resistance in C glabrata
Regulation of pleiotropic drug resistance in yeast
Regulation of multidrug resistance in pathogenic fungi
21 269 287 Effect of Niobium and Vanadium as an Alloying Elements in Tool Steels
discourse markers in writing
Molecular Recognition in Dendrimers
Piórkowska K Managers' loyalty as an organizational resource in the strategic context
Rueda Electromagnetic Zero Point Field as Active Energy Source in the Intergalactic Medium (1999)
Genolevures comparative genomics and molecular evolution of yeast
Molecular Gastronomy In The Uk
Proteomics of drug resistance in C glabrata
75 1067 1073 Elimination of Lubricants in Industries in Using Self Lubricating Wear Resistant
Ionic liquids as solvents for polymerization processes Progress and challenges Progress in Polymer
[US 2005] 6864611 Synchronous generator for service in wind power plants, as well as a wind power
więcej podobnych podstron