A Ser49Cys Variant in the Ataxia Telangiectasia,
Mutated, Gene that Is More Common in Patients with
Breast Carcinoma Compared with Population Controls
Thomas A. Buchholz,
M.D.
1
Michael M. Weil,
Ph.D.
2
Cheryl L. Ashorn,
M.S.
2
Eric A. Strom,
M.D.
1
Alice Sigurdson,
Ph.D.
3
Melissa Bondy,
Ph.D.
3
Ranajit Chakraborty,
Ph.D.
4
James D. Cox,
M.D.
1
Marsha D. McNeese,
M.D.
1
Michael D. Story,
Ph.D.
2
1
Department of Radiation Oncology, The Univer-
sity of Texas M. D. Anderson Cancer Center, Hous-
ton, Texas.
2
Department of Experimental Radiation Oncology,
The University of Texas M. D. Anderson Cancer
Center, Houston, Texas.
3
Department of Epidemiology, The University of
Texas M. D. Anderson Cancer Center, Houston,
Texas.
4
Center for Genome Information, Department of
Environmental Health, University of Cincinnati, Cin-
cinnati, Ohio.
Supported by Department of Defense Breast Can-
cer Research Program Career Development Award
BC980154 (T.A.B.) and by the Kleberg Fund for
New and Innovative Research (M.D.S.).
The authors thank Scott Manatt for his contribution
in collecting samples for the control population.
Dr. Buchholz, Dr. Weil, and Dr. Story contributed
equally to the current work.
Address for reprints: Thomas A. Buchholz, M.D.,
Department of Radiation Oncology, P.O. Box 97,
The University of Texas M. D. Anderson Cancer
Center, 1515 Holcombe Boulevard, Houston, TX
77030;
Fax:
(713)
563-2336;
E-mail:
tbuchhol@mdanderson.org
Received October 22, 2003; revision received De-
cember 24, 2003; accepted January 8, 2004.
BACKGROUND.
Mothers of children who have ataxia telangiectasia have been re-
ported to be at increased risk for development of breast carcinoma. To test whether
sequence variants in the ataxia telangiectasia, mutated, gene (ATM) are associated
with breast carcinoma, the authors compared the frequency of ATM cDNA se-
quence changes in patients with breast carcinoma with the corresponding fre-
quency in control patients.
METHODS.
The authors sequenced ATM cDNA from 91 patients with breast carci-
noma and compared the frequencies of sequence changes in these patients with
the corresponding frequencies in a control sample of 940 individuals with no
history of malignant disease.
RESULTS.
Thirty-five patients with breast carcinoma had one or more single-base
changes in ATM. Three genetic variants were found in at least two patients. These
variants resulted in Asp1853Asn, Pro1054Arg, or Ser49Cys amino acid substitutions
in the ATM protein. The Ser49Cys variant was more common in patients with
breast carcinoma than in the control patients, with respective frequencies of 6.7%
(5 of 75 patients) and 1.3% (12 of 940 patients; P
⫽ 0.006; Fisher two-sided exact
test). The subgroup of patients with bilateral breast carcinoma had a Ser49Cys
frequency of 11.8% (2 of 17 patients), which again was significantly different from
what was observed in the control group (P
⫽ 0.024; Fisher two-sided exact test).
The allele frequencies of the other two variants were not different between case
patients and control patients.
CONCLUSIONS.
Patients with breast carcinoma, particularly those with bilateral
disease, were more likely to have a variant in the ATM gene that resulted in a
Ser49Cys substitution in the gene product. Additional studies are needed to eval-
uate the potential functional consequences of the Ser49Cys substitution and con-
firm the relevance of this variant in the development of breast carcinoma. Cancer
2004;100:1345–51. © 2004 American Cancer Society.
KEYWORDS: breast carcinoma, ataxia telangiectasia, mutated, gene, variants, poly-
morphisms.
T
he genetic determinants of breast carcinoma risk remain elusive
for the majority of patients. However, recent evidence suggests
that abnormalities in the ataxia telangiectasia, mutated, gene (ATM)
may be a contributor to breast carcinoma risk. Individuals with ho-
mozygous ATM mutations develop the rare autosomal recessive dis-
ease ataxia telangiectasia (AT). A germline mutation in one ATM allele
(ATM heterozygosity) does not produce a recognizable phenotypic
change, but epidemiologic evidence suggests that ATM heterozygosity
may increase the relative risk of breast carcinoma.
1– 4
The first evi-
dence linking ATM heterozygosity and breast carcinoma was provided
1345
© 2004 American Cancer Society
DOI 10.1002/cncr.20133

by studies of families with AT. From these studies, it
was estimated that 6 – 8% of the breast carcinomas in
the United States occur in ATM heterozygotes,
1–5
which would make this condition a more common
genetic contributor to breast carcinoma than the com-
bined number of patients with a mutation in BRCA1 or
BRCA2.
The ATM gene was cloned in 1995. Since then, a
number of groups have undertaken molecular epide-
miology studies to test for an association between
ATM heterozygosity and the development of breast
carcinoma. These screening studies have produced
contradictory results, making interpretation diffi-
cult.
6 –15
Each study involved a different subset of pa-
tients with breast carcinoma, sample sizes were small,
and a variety of assays were used to detect ATM mu-
tations. The methodology used to detect ATM muta-
tions is of particular importance. Mutations in pa-
tients
with
AT
led
to
protein
truncations
in
approximately 80% of the case patients, which led
some investigators to use a protein truncation assay
for ATM heterozygosity studies. In general, the studies
using these assays have failed to find an association
between ATM mutations and breast carcinoma.
13,15
These studies suggest that deletions or major frame-
shift mutations in ATM are not significant contributors
to breast carcinoma risk. It is less clear whether some
ATM missense mutations are associated with a signif-
icant risk for the development of breast carcinoma.
Based on direct sequencing of the ATM gene in
patients with breast carcinoma, a number of studies
have reported that a significant percentage of patients
have single-base change variants.
6,9 –12,14
However, the
biologic significance of these changes remains un-
known, and it is unclear whether these variants di-
rectly contribute to breast carcinoma risk.
In the current study, our goal was to determine
the relevance of single-base changes to the risk of
breast carcinoma. We sequenced ATM cDNA samples
from patients with breast carcinoma and studied the
frequencies of identified variants in a large control
population. One of the variants that we identified was
significantly more common in patients with breast
carcinoma.
MATERIALS AND METHODS
Study Population
The current study was conducted using protocols ap-
proved by the surveillance committee of The Univer-
sity of Texas M. D. Anderson Cancer Center (Houston,
TX). Written informed consent was obtained from all
participants.
A total of 111 patients with breast carcinoma who
were recruited between 1997 and 2000 participated in
the study. In 20 patients, the sequencing of ATM was
unsuccessful, leaving 91 evaluable participants. Most
of the patients were enrolled at the time of radiation
treatments for breast carcinoma. Patients with breast
carcinoma who had previous histories of malignant
disease were specifically recruited for the study, al-
though all women with a history of breast carcinoma
were eligible.
Control Patients
Blood samples from 940 control patients were ob-
tained during institutionally sponsored, community-
based blood drives. Control participants did not have
a personal history of malignant disease. Information
on family history and other specific data were not
obtained.
Analysis of the ATM Gene
Consenting participants donated 20 mL of blood. A
specimen of total RNA was extracted from lympho-
cytes isolated by centrifugation on Ficoll-Hypaque
(Sigma Chemical Co., St. Louis, MO). Total lympho-
cyte RNA specimens were prepared according to the
method of Chomczynski and Sacchi
16
using commer-
cially available reagents (RNAzol B; Tel-Test, Friends-
wood, TX). Total RNA specimens were reverse-tran-
scribed and the ATM cDNA subsequently amplified via
the polymerase chain reaction (PCR). The PCR primer
sets were designed to amplify the ATM cDNA as eight
overlapping products ranging in size from 1200 to
1600 base pairs. Reverse-transcription PCR products
were purified by agarose gel electrophoresis and se-
quenced using commercially available cycle sequenc-
ing methodology and
33
P-labeled chain terminators
(Amersham, Piscataway, NJ). The PCR primers dou-
bled as sequencing primers. A Genomyx-LX sequenc-
ing apparatus (Genomyx Corp., Foster City, CA) was
used for electrophoretic resolution of the sequencing
products. Each reaction was assayed on two gels. The
first gel was designed to resolve from the primer to 350
bases, and the second was designed to resolve from
the primer to 800 bases. After electrophoresis, the
dried sequencing gel was exposed to X-ray film, and
the radiograph was analyzed for genetic variants in the
ATM cDNA. We defined ATM genetic variants as any
sequence other than the normal ATM sequence de-
fined in GenBank (reference sequence, HSU33841).
We confirmed the validity of our cDNA sequencing
assay via the analysis of two obligate AT heterozy-
gotes. Figure 1 shows a schematic display of the ATM
protein, and Table 1 describes the primers used in
sequencing.
1346
CANCER April 1, 2004 / Volume 100 / Number 7
Allele-Specific Oligonucleotide Assay
We used an allele-specific oligonucleotide (ASO) assay
to screen DNA samples from the control populations
for selected sequence variants originally identified in
patients with breast carcinoma. We did not sequence
the entire ATM gene in any of the control patients. The
result of the ASO assay was a simple plus/minus de-
termination of a single-base change at the location of
interest and a determination of whether one or both
alleles were affected.
The assay that we developed was based on the
assay designed by the DNA Diagnosis Laboratory at
the Baylor Institute for Molecular Genetics (Houston,
TX) to analyze the cystic fibrois gene.
17
Hybridization
probes were designed to avoid G:T or G:A mismatches
that would result in false-positive results. Ninety-six
samples were examined at a time, in paired blots. One
blot was tested for the correct nucleotide, and the
other blot was tested for the single-base change in
question. Each blot also contained positive and nega-
tive controls that were obtained from patient DNA
samples that may have contained the single-nucle-
otide change (as determined by sequencing). Each
time, the positive and negative controls matched the
results of the sequencing. Blot signal intensity was
quantified with storage phosphor technology. On the
rare occasion that the ASO results were ambiguous,
DNA from the sample was sequenced across the ATM
region of interest to determine nucleotide identity.
Statistical Methods
Any variant in ATM that occurred in at least two pa-
tients with breast carcinoma was of interest. An ASO
assay was developed for these variants and the fre-
quency of these variants was tested in the control
population. The frequencies of these variants were
then compared between case patients and control
patients. Because normal gene polymorphisms can
have different frequencies across ethnic groups, we
first compared the frequency of each variant across
the different ethnic groups in the control population.
If no difference was noted, the frequency of the par-
ticular genetic variant in the entire cohort of patients
was compared with that in the entire control popula-
tion. If a difference was noted across ethnic groups,
the frequency of the particular genetic variant in Cau-
casian case patients was compared with that in Cau-
casian control patients, because they constituted
the largest subgroup. For one identified variant
(Asp1853Asn), two individuals were homozygous for
the variant (Asn) allele. Therefore, for this variant, we
compared the allele frequencies between case patients
and control patients. All comparisons were made with
the Fisher exact tests. Two-sided P values
⬍ 0.05 were
considered significant.
RESULTS
Of the 111 patients with breast carcinoma enrolled in
the current study, the sequencing assay was unsuc-
cessful in 20 individuals. For 67 of the remaining 91
patients, the full length of the cDNA of the ATM gene
was successfully sequenced. One or more of the eight
overlapping regions of the cDNA were not successfully
sequenced in the remaining patients. The region not
sequenced varied among patients. No mutations that
would lead to protein truncation were identified.
However, it is possible that a truncation or deletion
mutation went undetected because of gel purification
of PCR products or nonsense-mediated decay of the
mRNA.
Table 2 displays the demographics and character-
istics of the patients with breast carcinoma. The me-
dian age of the participants was 55 years. The majority
(75%) of patients were Caucasian (all ethnic informa-
tion was self-reported). Forty-nine of the 91 patients
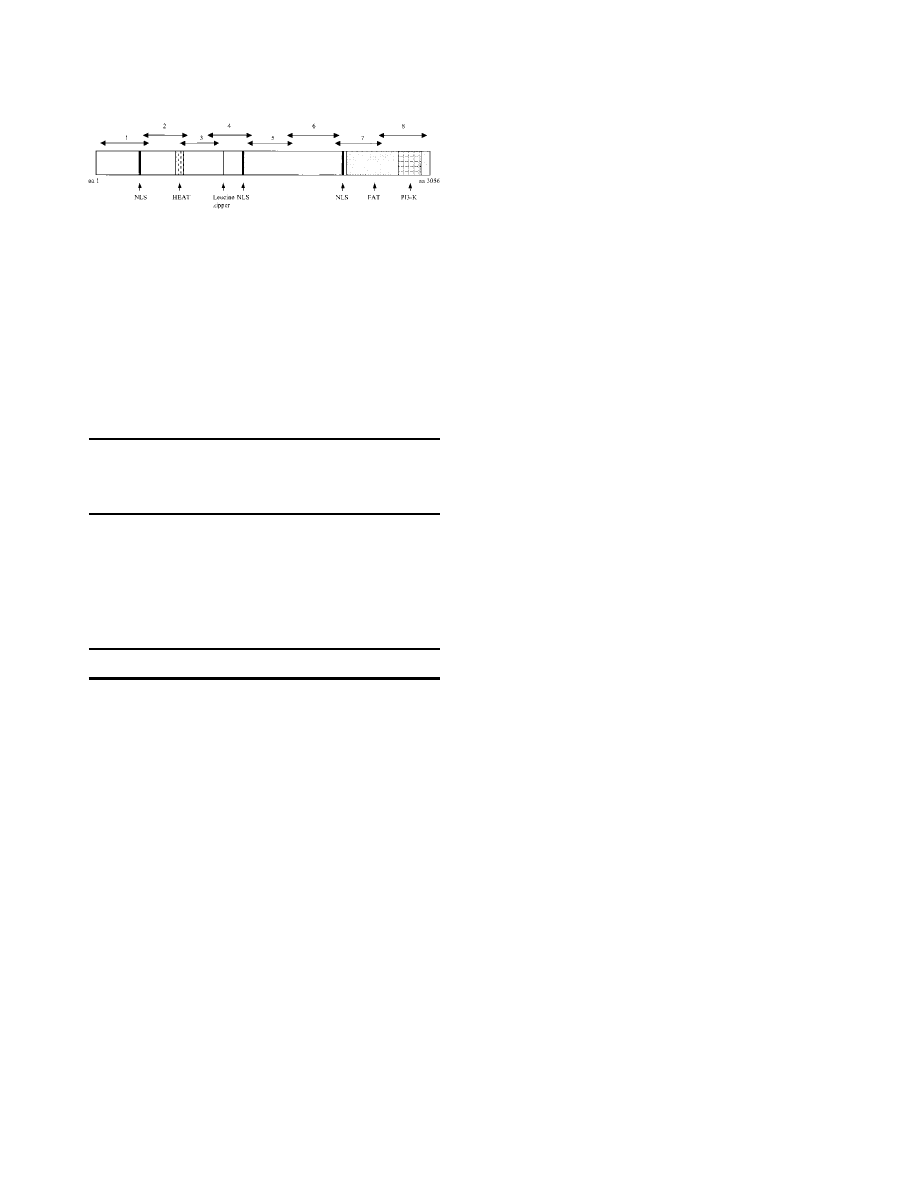
FIGURE 1.
Schematic display of representative domains of the ataxia
telangiectasia, mutated, protein (ATM). Domains are listed as follows: NLS,
nuclear localization sequence; HEAT, sequence elements conserved in Hun-
tington protein (H), elongation factor 3 protein (E), protein phosphatase 2a
protein (A), and Tor 1p protein (T); FAT, conserved region in FRAP (F), ATM (A),
and TRRAP (T) proteins, including a highly conserved 30 –amino acid residue
tail; and PI3-K, the C-terminal kinase. Also shown is the leucine zipper region.
Horizontal arrows indicate overlapping regions of cDNA sequenced as outlined
in Table 1. aa: amino acid.
TABLE 1
Overlapping Regions of cDNA Sequenced
Region
no.
Primer pair
ATM cDNA
region
sequenced
(nt)
1
5
⬘tgaaattgtgaaccatgagtc 3⬘ttggggtagaagctgagatag 177–1504
2
5
⬘gcaaaaggaagaaaatagaac 3⬘ctcaagcaacgtgtacatagc 1340–2532
3
5
⬘ctgttacatgggtgtaatagc3 3⬘atccaaagtttcagggttctc 2376–3604
4
5
⬘ggctgcagagtcaatcaatag 3⬘ggagaagctacgtaatgacac 3453–4645
5
5
⬘taaaaagtggcttaggaggag 3⬘aacatgtgtagaaagcagatt 4547–5767
6
5
⬘agttcgatcagcagctgttac 3⬘ttcagagagttgtctatgtgt 5373–6768
7
5
⬘cagccttgagtctgtgtattc 3⬘tttaggcacatttttagttat 6672–7936
8
5
⬘gtttattatactggccttagc 3⬘tgagatttttggggtctatgg 7860–9305
ATM: ataxia telangiectasia, mutated; nt: nucleotides.
Genetic Variant in ATM and Breast Carcinoma/Buchholz et al.
1347
had a history of a second malignancy independent of
their breast carcinoma, the most common type being
a second breast carcinoma (25%). Other types in-
cluded lymphoma, soft tissue sarcoma, leukemia, mel-
anoma, and endometrial, anal, colon, thyroid, lung,
ovarian, and gastric carcinoma. Normal tissue injuries
from treatment were recorded for patients treated
with radiation. Among the control patients, 56% were
Caucasian, 23% were Hispanic American, 15% were
African American, 4% were Asian American, and 2%
were of another ethnicity (all ethnic information was
self reported). In addition, 52% were women, and 48%
were men.
Several single-base change variants were identi-
fied among patients with breast carcinoma. Fifty-six
patients (62%) had the reference sequence. Twenty-
four (26%) had one sequence variant indentified, and
the remaining 11 (12%) had two or more variants.
Four genetic variants were found in three or
more case patients; these variants were 5557G
3A,
3161C
3G, 145C3G, and 4578C3T, which caused the
following amino acid substitutions in the ATM
protein: Asp1853Asn, Pro1054Arg, Ser49Cys, and
Pro1526Pro, respectively. The Asp1853Asn substitu-
tion was the most common variant, with two homozy-
gotes and 17 heterozygotes for the variant (Asn) allele.
Neither of the homozygote patients had been diag-
nosed with AT or had clinical findings associated with
the disease. Ser49Cys was the next most common
variant, followed by the Pro1054Arg and Pro1526Pro
variants. No patient was homozygous for these three
variants. Two additional variants, Asp126Glu and
Asp1853Val, were found in two case patients.
The development of the ASO assay was successful
for the Asp1853Asn, Pro1054Arg, and Ser49Cys variants,
but not for the Pro1526Pro variant. Table 3 shows the
allele frequency of the three investigated genetic variants
in the control patients and the inferred haplotypes and
their frequencies classified by ethnicity. Within the con-
trol population, there were ethnic differences in the
allele frequencies at the Asp1853Asn and Pro1054Arg
sites. Therefore, case– control frequency comparsions
were performed using only Caucasian individuals. In
contrast, no ethnic or gender-based difference in allele
frequencies was detected for the Ser49Cys site. Hence,
the freqeuency of the variant at this site was compared
across the entire set of control samples.
Table 4 shows the comparison of the frequency of
these genetic variants between case patients and con-
trol patients. There were no statistical differences in
frequencies for the Asp1853Asn and Pro1054Arg vari-
ants. However, the Ser49Cys variant was found in 6.7%
(5 of 75) of patients with breast carcinoma, compared
with 1.3% (12 of 940) of patients in the control group
(P
⫽ 0.006; Fisher two-sided exact test). Individuals
carrying the Cys variant allele at this site had a 5.52-
fold greater risk of developing breast carcinoma com-
pared with patients who were homozygous for the
Ser/Ser allele (odds ratio [OR], 5.52; 95% confidence
interval [CI], 1.89 –16.12). It is noteworthy that a fre-
quency of 11.8% (2 of 17 patients) was observed in the
subgroup of patients with bilateral breast carcinoma;
again, this frequency was significantly different from
the frequency observed in the control group (P
⫽ 0.025; Fisher two-sided exact test). Only one of the
five patients with breast carcinoma who had the
Ser49Cys variant had a family history of breast carci-
noma. The OR for bilateral breast carcinoma for the
Cys variant– carrying individuals was 10.31 (95% CI,
2.12–50.13) relative to individuals who were homozy-
gous for Ser/Ser. Nine patients developed a clinically
relevant complication after radiation treatment, and
none of these patients had the Ser49Cys variant.
DISCUSSION
In the current study, we identified a variant in the
ATM gene that resulted in a Ser49Cys substitution in
the gene product. This specific single-nucleotide
TABLE 2
Characteristics of the Patients with Breast Carcinoma
Characteristic
No. (%)
Age (yrs)
⬍ 40
10 (11)
40–49
19 (21)
⬎ 50
61 (68)
Ethnicity
Caucasian
68 (75)
Hispanic American
10 (11)
African American
8 (9)
Asian American
3 (3)
Other
2 (2)
History of second malignancy
None
42 (46)
Second breast
23 (25)
Other
26 (29)
Family history of breast carcinoma
First-degree relative
68 (75)
Other relative
10 (11)
Negative
8 (9)
Unknown
3 (3)
Stage (AJCC)
Ductal carcinoma in situ
4 (4)
I
32 (31)
II
45 (44)
III
22 (21)
Type of surgery
Breast conservation
38 (38)
Mastectomy
63 (62)
AJCC: American Joint Committee on Cancer.
1348
CANCER April 1, 2004 / Volume 100 / Number 7
change was more common among patients with
breast carcinoma compared with control patients.
Many other investigators have suggested that single-
base changes in the ATM gene may be associated with
an increase in breast carcinoma risk. However, to our
knowledge, the current report is the first to link breast
carcinoma to the Ser49Cys genetic variant. The sam-
ple size was relatively small, and therefore, the find-
ings of the current study should be considered pre-
liminary, hypothesis-generating data that require
validation. However, one strength of the current study
was the large control group, which enabled us to de-
termine the allelic frequencies of these variants in a
control population. For example, two of the three ge-
netic variants that were present in two or more case
patients were equally represented in the control pop-
ulation.
The interest in the relation between ATM and
breast carcinoma began after the observation that
mothers and grandmothers of children with AT (obli-
gate heterozygotes) had a 5.1-fold increased risk of
developing breast carcinoma.
3
This original study
3
predated the cloning of the ATM gene, but the obser-
vation was reconfirmed in a study of 50 Nordic fami-
lies with AT.
4
In that study, the standard incidence
ratio for the development of breast carcinoma was 7.1
in obligate heterozygotes (95% CI, 2.3–17). In addition,
other studies have confirmed the increased breast car-
cinoma risk in obligate heterozygote individuals.
5
The cloning of the ATM gene not only allows gen-
otype-based assays of patients with breast carcinoma,
such as the one we report, but also permits new lab-
oratory methods to investigate the relation between
ATM and breast carcinoma. For example, a recent
study found that mice genetically engineered to be
heterozygous for a 7636del9 truncating mutation in
ATM developed malignancies at an increased fre-
quency, probably due to the production of an abnor-
TABLE 3
Allele Frequency of Genetic Variants in the Control Population
Variant
Caucasian
(%)
African
American
(%)
Hispanic
American
(%)
Asian
American
(%)
Other
(%)
Homogeneity
(P value)
Asp1853Asn
⬍ 0.00005
Wild-type
907 (86)
278 (98)
396 (93)
83 (99)
32 (100)
Variant
149 (14)
6 (2)
28 (7)
1 (1)
0 (0)
Pro1054Arg
0.0473
Wild-type
1021 (97)
282 (99)
416 (98)
84 (100)
31 (97)
Variant
35 (3)
2 (1)
8 (2)
0 (0)
1 (3)
Ser49Cys
0.3634
Wild-type
1046 (99)
284 (100)
422 (99.5)
84 (100)
32 (100)
Variant
10 (1)
0 (0)
2 (0.5)
0 (0)
0 (0)
TABLE 4
Frequency of Genetic Variants in Case and Control Patients
Site
Frequency (%)
Allele frequencies (wild-type/variant)
P value
Cases
Controls
Cases
Controls
Asp1853Asn
a
Asp/Asp
39/58 (67.2)
394/528 (74.6)
0.818/0.181
0.8589/0.1411
0.807
Asp/Asn
17/58 (29.3)
119/528 (22.5)
Asn/Asn
2/58 (3.4)
15/528 (2.8)
Pro1954Arg
a
Pro/Pro
58/61 (95.1)
476/510 (93.2)
0.9754/0.0246
0.9657/0.0343
0.841
Pro/Arg
3/61 (4.9)
33/510 (6.5)
Arg/Arg
0/61 (0)
1/510 (0.2)
Ser49Cys
Ser/Ser
70/75 (93.3)
928/940 (98.7)
0.9667/0.0333
0.9936/0.0064
0.006
Ser/Cys
5/75 (6.7)
12/940 (1.3)
Cys/Cys
0/75 (0)
0/940 (0)
a
Frequencies only in Caucasian case and control patients.
Genetic Variant in ATM and Breast Carcinoma/Buchholz et al.
1349
mal protein that acts as a dominant negative.
18
In
contrast, knockout heterozygous ATM mice did not
have an increased susceptibility.
18
This same study
also found evidence of an association between the
7636del9 mutation and the development of breast car-
cinoma in humans.
18
These data suggest that not all
ATM mutations will have the same affect on cellular
phenotype and the risk of cancer development. Spe-
cifically, for ATM heterozygosity to affect cancer risk,
the specific mutation must produce a dominant-neg-
ative protein product, a hypothesis that is further sup-
ported by other preclinical work.
19
However, this hy-
pothesis is inconsistent with the aforementioned
epidemiologic data indicating that obligate heterozy-
gote individuals, who most often have a mutation that
truncates the ATM protein, have an increased breast
carcinoma risk.
The production of a dominant-negative protein
may explain why the data linking genetic changes in
ATM to cancer risk have been inconsistent. Table 5
provides an overview of the studies that investigated
the relation between ATM genetic variants and the risk
of developing breast carcinoma.
6 –15
A number of stud-
ies have suggested that single-base oligonucleotide
changes may increase the risk of breast carcinoma
development, but further laboratory studies are
needed to clearly show that these variants lead to a
change in cellular phenotype. No previous published
reports
have
investigated
whether
the
specific
Ser49Cys variant affects cellular phenotype. Further-
more, the Ser49Cys position in the ATM protein is not
recognized as being part of a structural or functional
protein domain.
The novel findings in the current study suggesting
a relation between the Ser49Cys variant in ATM and
breast carcinoma risk require independent validation.
Our case population size was relatively small, and only
five case patients had this sequence variant. Nonethe-
less, the difference in the frequency of this variation in
case patients versus control patients was highly signif-
icant. The higher frequency of the Ser49Cys variant in
women with bilateral breast carcinoma adds support
to the proposal that the identity of the variant affects
risk.
The Ser49Cys variant has been reported by other
authors, although the current report is the first to find
TABLE 5
Series Investigating the Role of the ATM Gene in Breast Carcinoma
First author
Method
Case sample
size
Control group
Conclusions
Thorstenson et al., 2003
6
DNA sequencing
270
Yes (n
⫽ 122)
L1420F variant more common in high-risk population;
other variants also identified
Offit et al., 2002
7
cDNA sequencing
37
No
ATM probably not a factor in breast carcinoma
development after radiation for Hodgkin Disease
Chenevix-Trench et al., 2002
8
Mutation-specific
assay
525 or 262
a
Yes (n
⫽ 381 or 68)
a
T7271G and IVS10-6T
3 G variants more common in
familial breast carcinoma
Sommer et al., 2002
9
DNA sequencing
43
Yes (n
⫽ 43)
A variety of single-base changes more common in
patients with breast carcinoma; specific changes not
compared
Do¨rk et al., 2001
10
DNA sequencing/
mutation-
specific assay
192/1000
b
Yes (n
⫽ 500)
T7271G and IVS10-6T
3 G variants more common in
patients with breast carcinoma
Teraoka et al., 2001
11
cDNA sequencing
258
Yes (n
⫽ 81)
Single-base changes more common in case patients with
young age or positive family history
Broeks et al., 2000
12
DNA sequencing
82
No
Single-base changes, including IVS10-6T
3 G, may
contribute to breast carcinoma development
Shafman et al., 2000
13
cDNA truncation
assay
57
No
Truncation mutations do not contribute to the incidence
of a second breast carcinoma after radiation for first
breast carcinoma
Izatt et al., 1999
14
DNA sequencing
100
Yes (n
⫽ 106)
Germline mutations rare in patients with breast
carcinoma age
⬍ 40 yrs
FitzGerald et al., 1997
15
Protein truncation
assay
400
Yes (n
⫽ 202)
Truncation mutations not associated with breast
carcinoma risk
Current series
cDNA sequencing
91
Yes (n
⫽ 940)
Ser49Cys more common in patients with breast
carcinoma
ATM: ataxia telangiectasia, mutated.
a
The T7271G variant was examined in 525 case patients and 381 control patients, and the IVS10-6T
3 G 525 variant was examined in 262 case patients and 68 control patients.
b
One hundred ninety-two case patients had sequencing of the ATM gene, and 1000 case patients had only the region of the gene that was specific to the variants examined.
1350
CANCER April 1, 2004 / Volume 100 / Number 7

this variant more often in case patients than in control
patients. Vorechovsky et al.
20
were the first to report
this variant and considered it to be a rare polymor-
phism, present in only 1 of 49 breast carcinoma tu-
mors or cell lines. Subsequently, Izatt et al.
14
reported
this variant in 1 of 100 patients age
⬍ 40 years with
breast carcinoma, compared with 1 of 50 control pa-
tients. Finally, Do¨rk et al.
10
found the Ser49Cys variant
in 3 of 192 unselected breast carcinoma cases. It is
unclear as to why we found a higher frequency of the
Ser49Cys variant in the current patient population
compared with previous reports. This discrepancy
may be attributable to differences in the study popu-
lation. For example, Do¨rk et al. studied a predomi-
nantly German population; in addition, the current
population had a higher percentage of patients with
bilateral breast carcinoma, and the Ser49Cys variant
was observed in 11% of this subgroup. Of course, the
difference may also be due to chance.
One method for validating the relevance of the
Ser49Cys variant to breast carcinoma development
would involve testing for this specific variant in a large
independent data set. This methodology has been
used previously for other genetic variants in ATM. For
example, after Broeks et al.
12
reported in a relatively
small study that the IVS10-6T
3G variant may contrib-
ute to the development of breast carcinoma, Chene-
vix-Trench et al.
8
tested specifically for this variant
(without complete gene sequencing) in a much larger
case– control study. They confirmed that this variant
was overrepresented in families with multiple breast
carcinomas. A second method for testing the rele-
vance of single-base changes found in association
studies involves genetically engineering cells or mice
to carry the relevant genetic variant. This method,
which has been successfully used to evaluate other
single-base variants of ATM,
18
allows one to test
whether the specific change leads to the production of
a dominant-negative protein that can affect cellular
phenotype.
In conclusion, we found that a single-base genetic
variant in the ATM gene that leads to a Ser49Cys
change in the protein product was statistically over-
represented in a breast carcinoma population com-
pared with a control population. These results are
useful as hypothesis-generating data that justify fur-
ther investigation aimed at determining whether this
variant may confer an increased risk of breast carci-
noma development.
REFERENCES
1.
Pippard EC, Hall AJ, Barker DJ, et al. Cancer in homozygotes
and heterozygotes of ataxia-telangiectasia and xeroderma
pigmentosum in Britain. Cancer Res. 1988;48:2929 –2932.
2.
Swift M, Reitnauer P, Morrell D, et al. Breast and other
cancers in families with ataxia telangiectasia. N Engl J Med.
1987;316:1289 –1294.
3.
Swift M, Morrell D, Massey R, et al. Incidence of cancers in
161 families affected by ataxia telangiectasia. N Engl J Med.
1991;325:1831–1836.
4.
Olsen JH, Hahnemann JM, Borresen-Dale AL, et al. Cancer
in patients with ataxia-telangiectasia and in their relatives in
the Nordic countries. J Natl Cancer Inst. 200;93:121–127.
5.
Morrell D, Chase CL, Swift M. Cancers in 44 families with
ataxia-telangiectasia. Cancer Genet Cytogenet. 1990;50:119 –
123.
6.
Thorstenson YR, Roxas A, Kroiss R, et al. Contributions of
ATM mutations to familial breast and ovarian cancer. Can-
cer Res. 2003;63:3325–3333.
7.
Offit K, Gilad S, Paglin S, et al. Rare variants of ATM and risk
for Hodgkin’s disease and radiation-associated breast can-
cers. Clin Cancer Res. 2002;8:3813–3819.
8.
Chenevix-Trench G, Spurdle AB, Gatei M, et al. Dominant
negative ATM mutations in breast cancer families. J Natl
Cancer Inst. 2002;94:205–215.
9.
Sommer SS, Buzin CH, Jung M, et al. Elevated frequency of
ATM gene missense mutations in breast cancer relative to
ethnically matched controls. Cancer Genet Cytogenet. 2002;
134:25–32.
10. Do¨rk T, Bendix R, Bremer M, et al. Spectrum of ATM gene
mutations in a hospital-based series of unselected breast
cancer patients. Cancer Res. 2001;61:7608 –7615.
11. Teraoka SN, Malone KE, Doody DR, et al. Increased fre-
quency of ATM mutations in breast carcinoma patients with
early onset disease and positive family history. Cancer. 2001;
92:479 – 487.
12. Broeks A, Urbanus JH, Floore AN, et al. ATM-heterozygous
germline mutations contribute to breast cancer-susceptibil-
ity. Am J Hum Genet. 2000;66:494 –500.
13. Shafman TD, Levitz S, Nixon AJ, et al. Prevalence of germline
truncating mutations in ATM in women with a second
breast cancer after radiation therapy for a contralateral tu-
mor. Genes Chromosomes Cancer. 2000;27:124 –129.
14. Izatt L, Greenman J, Hodgson S, et al. Identification of
germline missense mutations and rare allelic variants in the
ATM gene in early-onset breast cancer. Genes Chromosomes
Cancer. 1999;26:286 –294.
15. FitzGerald MG, Bean JM, Hegde SR, et al. Heterozygous
ATM mutations do not contribute to early onset of breast
cancer. Nat Genet. 1997;15:307–310.
16. Chomczynski P, Sacchi N. Single-step method of RNA iso-
lation by acid guanidinium thiocyanate-phenol-chloroform
extraction. Anal Biochem. 1987;162:156 –159.
17. DeMarchi JM, Richards CS, Fenwick RG, et al. A robotics-
assisted procedure for large scale cystic fibrosis mutation
analysis. Hum Mutat. 1994;4:281–290.
18. Spring K, Ahangari F, Scott SP, et al. Mice heterozygous for
mutation in Atm, the gene involved in ataxia-telangiectasia,
have heightened susceptibility to cancer. Nat Genet. 2002;
32:185–190.
19. Scott SP, Bendix R, Chen P, et al. Missense mutations but
not allelic variants alter the function of ATM by dominant
interference in patients with breast cancer. Proc Natl Acad
Sci U S A. 2002;99:925–930.
20. Vorechovsky I, Rasio D, Luo L, et al. The ATM gene and
susceptibility to breast cancer: analysis of 38 breast tu-
mors reveals no evidence for mutation. Cancer Res. 1996;
56:2726 –2732.
Genetic Variant in ATM and Breast Carcinoma/Buchholz et al.
1351
Wyszukiwarka
Podobne podstrony:
Functional and Computational Assessment of Missense Variants in the Ataxia Telangiectasia Mutated (A
Proton Magnetic Resonance Spectroscopy of the Medial Prefrontal Cortex in Patients With Deficit Schi
The Effects of Probiotic Supplementation on Markers of Blood Lipids, and Blood Pressure in Patients
Effect of high dose intravenous ascorbic acid on the level of inflammation in patients with rheumato
The relationship of Lumbar Flexion to disability in patients with low back pain
Effects of Clopidogrel?ded to Aspirin in Patients with Recent Lacunar Stroke
Difficult airway management in a patient with traumatic asphyxia
Impaired Sexual Function in Patients with BPD is Determined by History of Sexual Abuse
Konstatinos A Land versus water exercise in patients with coronary
Muscle Mass Gain Observed in Patients with Short Bowel Syndrome
Difficult airway management in a patient with traumatic asphyxia
Serum cytokine levels in patients with chronic low back pain due to herniated disc
Personality Constellations in Patients With a History of Childhood Sexual Abuse
Evaluation of the role of Finnish ataxia telangiectasia mutations in hereditary predisposition to br
Variants in the ATM gene associated with a reduced risk of contralateral breast cancer
ATM Gene Founder Haplotypes and Associated Mutations in Polish Families with Ataxia Telangiectasia
Variants in the ATM gene and breast cancer susceptibility
Jacobsson G A Rare Variant of the Name of Smolensk in Old Russian 1964
Jacobsson G A Rare Variant of the Name of Smolensk in Old Russian 1964
więcej podobnych podstron