Chapter 5: Gene Expression - Transcription
When a protein is needed by a cell, the genetic code for that protein
must be read from the DNA and processed.
A two step process:
1.
Transcription = synthesis of a single-stranded RNA molecule
using the DNA template (1 strand of DNA is transcribed).
2.
Translation = conversion of a messenger RNA sequence into the
amino acid sequence of a polypeptide (i.e., protein synthesis)
Both processes occur throughout the cell cycle. Transcription
occurs in the nucleus, whereas translation occurs in the
cytoplasm.
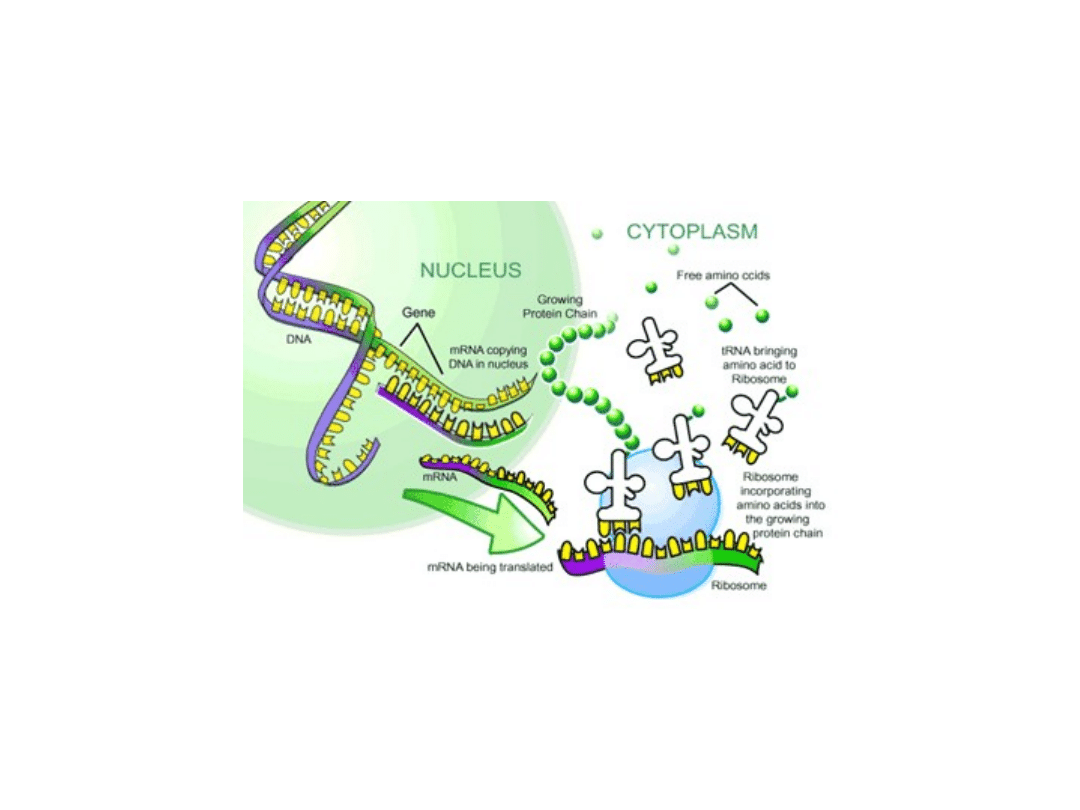
Five different types of RNA, each encoded by different genes:
1. mRNA
Messenger RNA, encodes the amino acid sequence
of a polypeptide.
2. tRNA
Transfer RNA, transports amino acids to ribosomes
during translation.
3. rRNA
Ribosomal RNA, forms complexes called ribosomes
with protein, the structure on which mRNA is
translated.
4. SnRNA
5. miRNA/siRNA
Small nuclear RNA, forms complexes with proteins
used in eukaryotic RNA processing (e.g., exon
splicing and intron removal).
Micro RNA/small interfering RNA, short ~22 nt RNA
sequences that bind to 3’ UTR target mRNAs and
result in silencing.

Transcription: How is an RNA strand synthesized?
1.
Regulated by gene regulatory elements within each gene.
2.
DNA unwinds next to a gene.
3.
RNA is transcribed 5’ to 3’ from the template (3’ to 5’).
4.
Similar to DNA synthesis, except:
NTPs instead of dNTPs (no deoxy-)
No primer
No proofreading
Adds Uracil (U) instead of thymine (T)
RNA polymerase
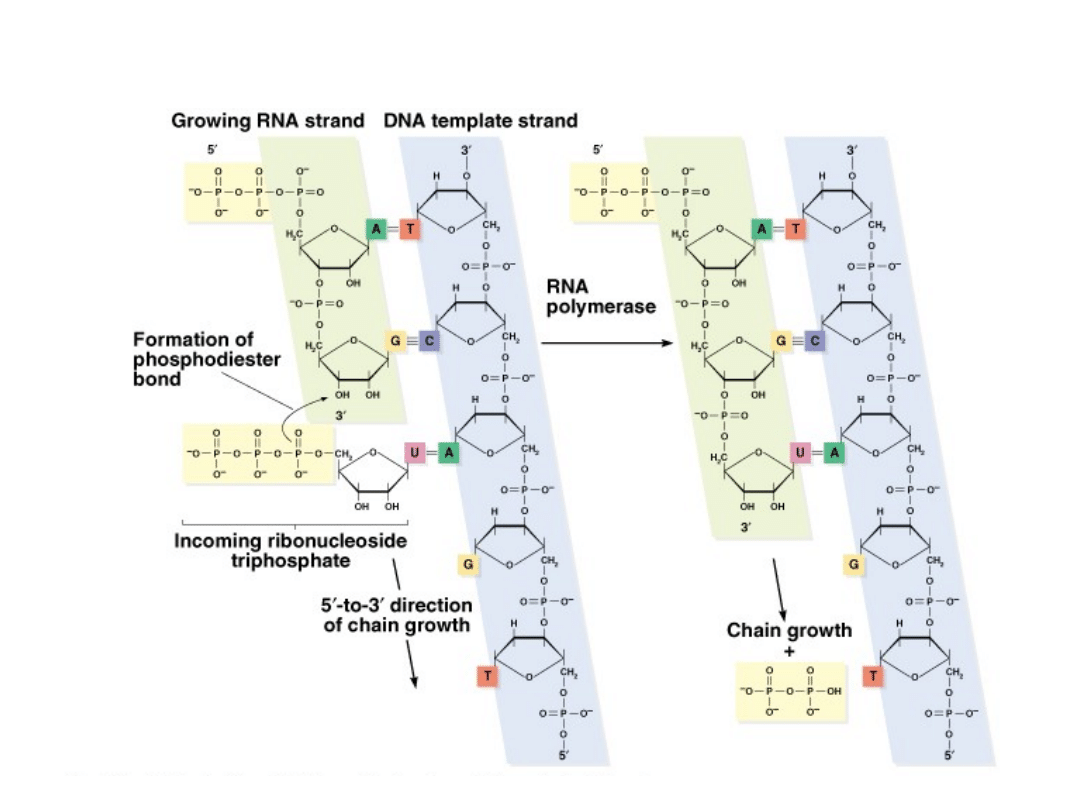
Fig. 5.2

Animation RNA biosynthesis

Three Steps to Transcription:
1.
Initiation
2.
Elongation
3.
Termination
Occur in both prokaryotes and eukaryotes.
Elongation is conserved in prokaryotes and eukaryotes.
Initiation and termination proceed differently.
Step 1-Initiation, E. coli model:
Fig. 5.3
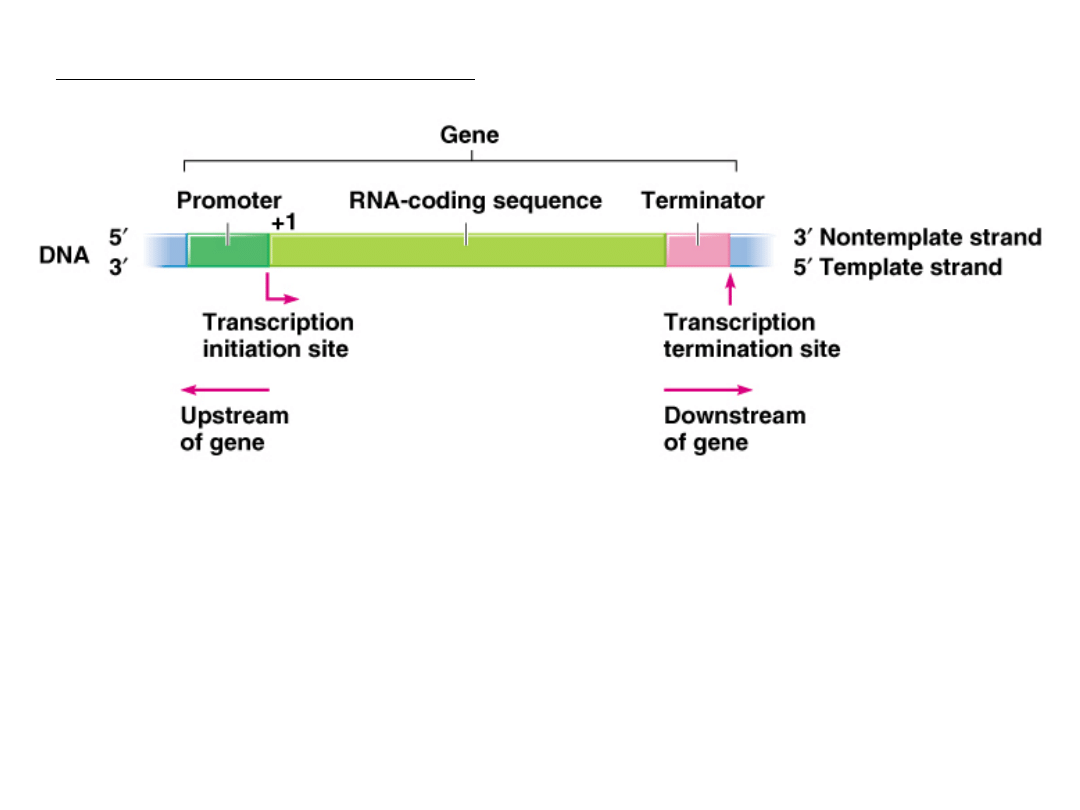
Each gene has three regions:
1.
5’ Promoter, attracts RNA polymerase
-10 bp 5’-TATAAT-3’
-35 bp 5’-TTGACA-3’
2.
Transcribed sequence (transcript) or RNA coding sequence
3.
3’ Terminator, signals the stop point

Step 1-Initiation
1.
RNA polymerase combines with sigma factor (a polypeptide) to
create RNA polymerase holoenzyme
Recognizes promoters and initiates transcription.
Sigma factor required for efficient binding and transcription.
Different sigma factors recognize different promoter
sequences.
2.
RNA polymerase holoenzyme binds promoters and untwists DNA
Binds loosely to the -35 promoter (DNA is d.s.)
Binds tightly to the -10 promoter and untwists
3.
Different types and levels of sigma factors influence the level and
dynamics of gene expression (how much and efficiency).
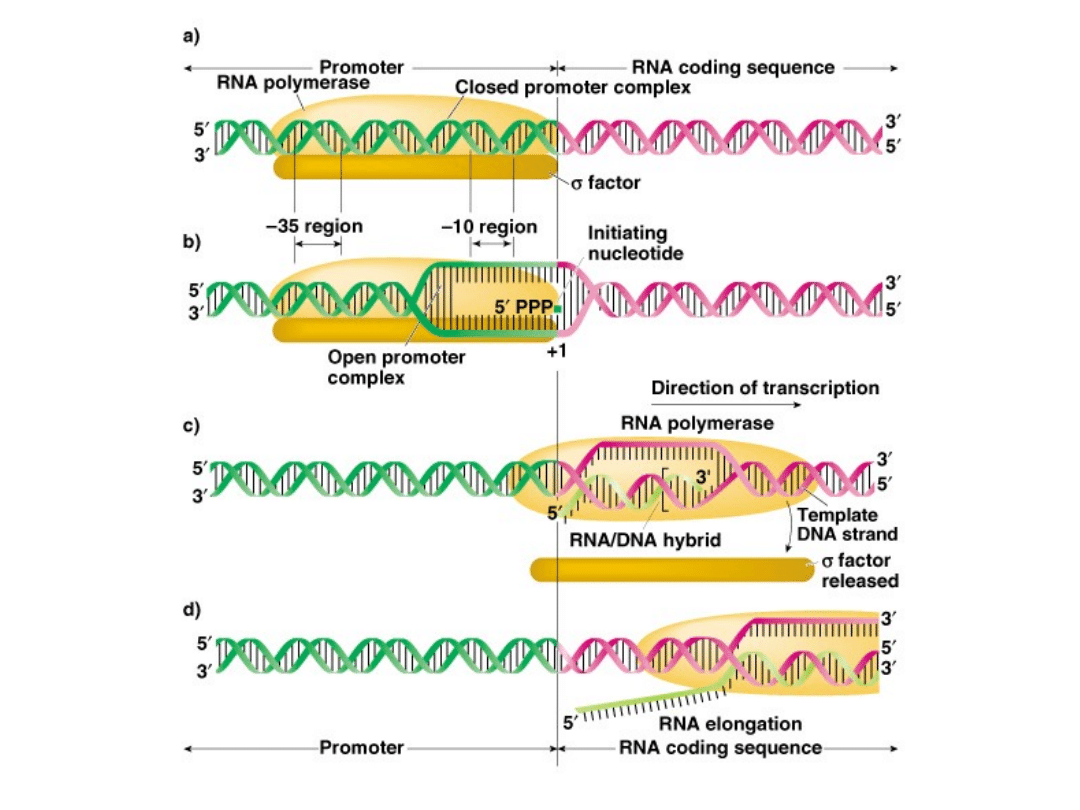
Fig. 5.4

Step 2-Elongation
1.
After 8-9 bp of RNA synthesis occurs, sigma factor is released
and recycled for other reactions.
2.
RNA polymerase completes the transcription at 30-50 bp/second.
3.
DNA untwists rapidly, and re-anneals behind the enzyme.
4.
Part of the new RNA strand is hybrid DNA-RNA, but most RNA is
displaced as the helix reforms.
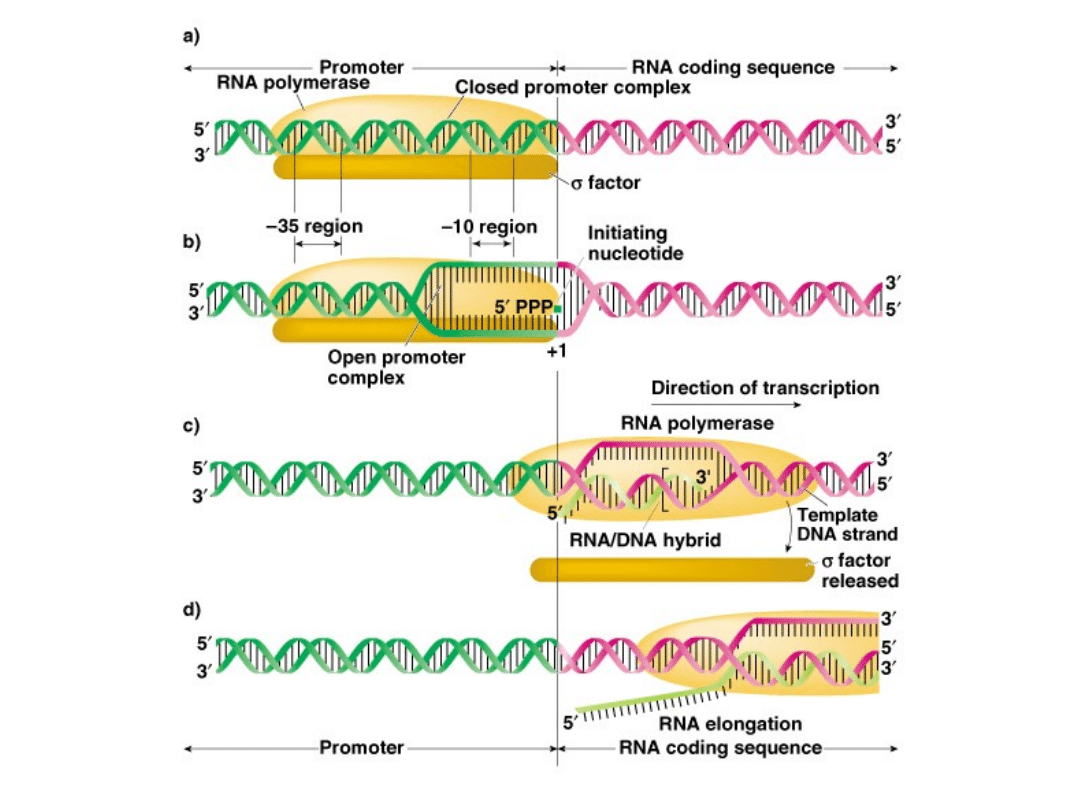
Fig. 5.4
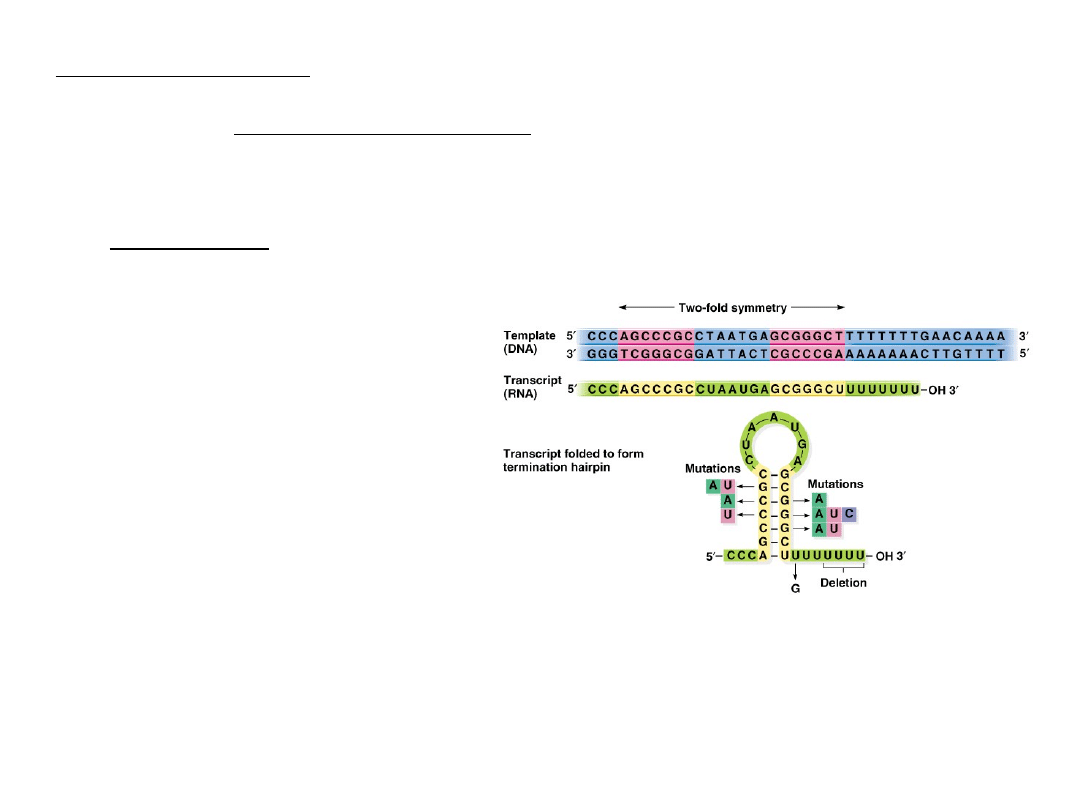
Step 3-Termination
Two types of terminator sequences occur in prokaryotes:
1.
Type I (-independent)
Palindromic, inverse repeat forms a hairpin loop and is believed
to physically destabilize the DNA-RNA hybrid.
2.
Type II (-dependent)
Involves factor proteins, believed to break the hydrogen bonds
between the template DNA and RNA.
Fig. 5.5
Prokaryotes possess only one type of RNA polymerase
transcribes mRNAs, tRNAs, and rRNAs
Transcription is more complicated in eukaryotes
Eukaryotes possess three RNA polymerases:
1.
RNA polymerase I, transcribes three major rRNAs 12S, 18S, 5.8S
2.
RNA polymerase II, transcribes mRNAs and some snRNAs
3.
RNA polymerase III, transcribes tRNAs, 5S rRNA, and snRNAs
*S values of rRNAs refer to molecular size, as determined by sucrose
gradient centrifugation. RNAs with larger S values are
larger/have a greater density.
Transcription of protein-coding genes by RNA polymerase II
RNA polymerase II transcribes a precursor-mRNA
We can divide eukaryotes promoter into two regions:
1.
The core promoters elements. The best characterized are
A short sequence called Inr (Initiator)
TATA Box = TATAAAA, located at about position -30
*AT-rich DNA is easier to denature than GC-rich DNA
2.
Promoter proximal elements (located upstream, ~-50 to
-200 bp)
“Cat Box” = CAAT and “GC Box” GGGCGG
Different combinations occur near different genes.
Transcription regulatory proteins (activators) and enhancers also
are required.
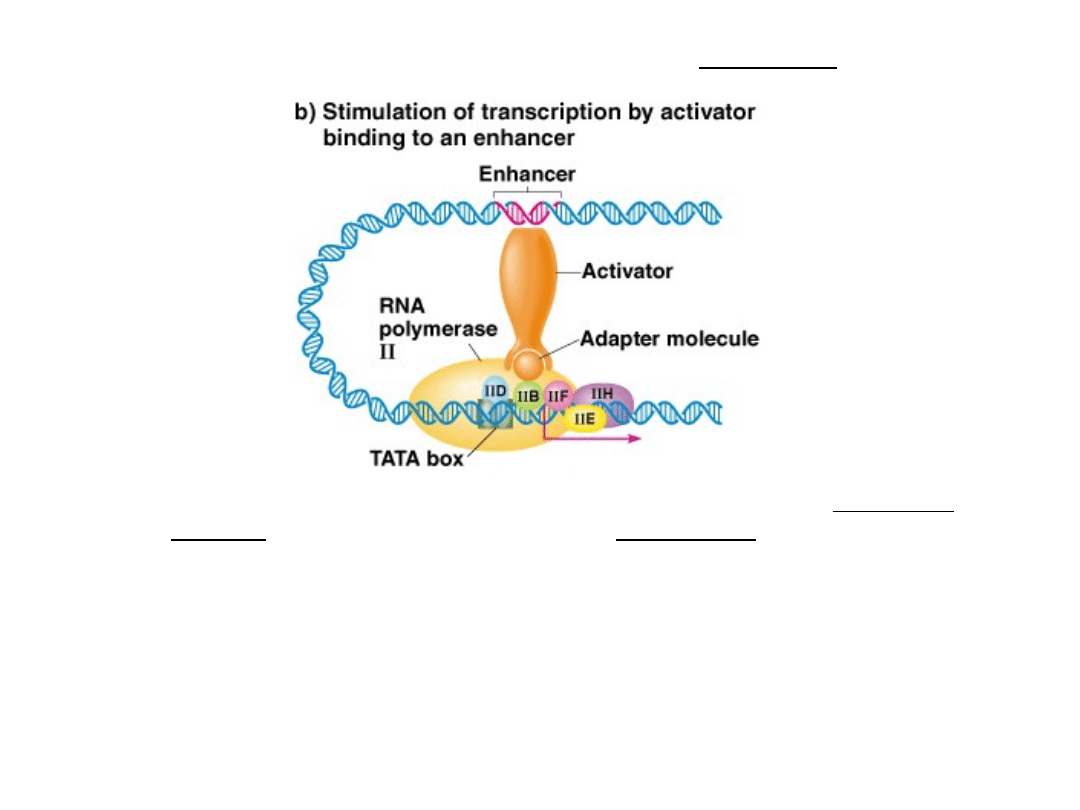
Transcription regulatory proteins = Activators
High-level transcription is induced by binding of activator
factors to DNA sequences called enhancers.
Enhancers are usually located upstream of the gene they
control, they modulate transcription from a distance.
Can be several kb from the gene
Silencer elements and repressor factors also exist

Transcription of protein-coding genes by RNA polymerase II
General Transcription factors (GTFs) also are required by RNA
polymerases (function is similar to sigma factor).
GTFs are proteins, assembled on the core promoter
Each GTF works with only one kind of RNA polymerase (required
by all 3 RNA polymerases).
Numbered (i.e., named) to match their RNA polymerase.
TFIID, TFIIB, TFIIF, TFIIE, TFIIH
Binding of GTFs and RNA polymerase occurs in a set order in
protein coding genes.
Complete complex (RNA polymerase + GTFs) is called a pre-
initiation complex (PIC).
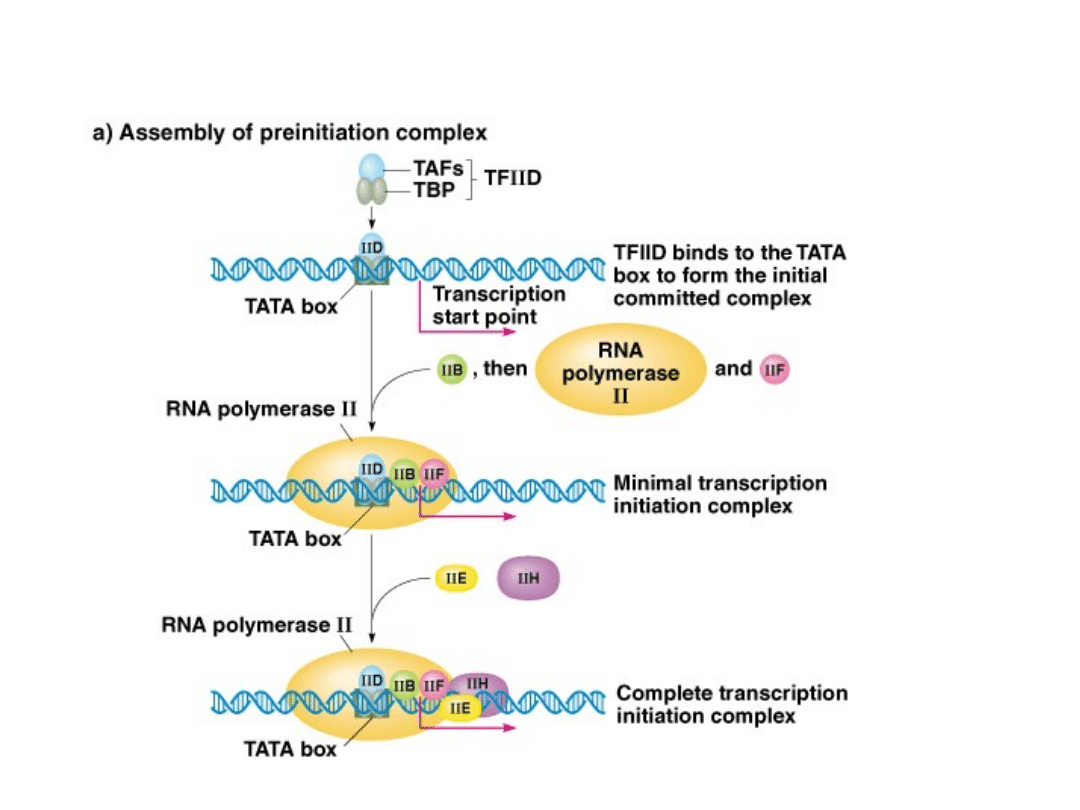
Order of binding is: IID + IIA + IIB + RNA poly. II + IIF +IIE +IIH
Fig. 5.7.
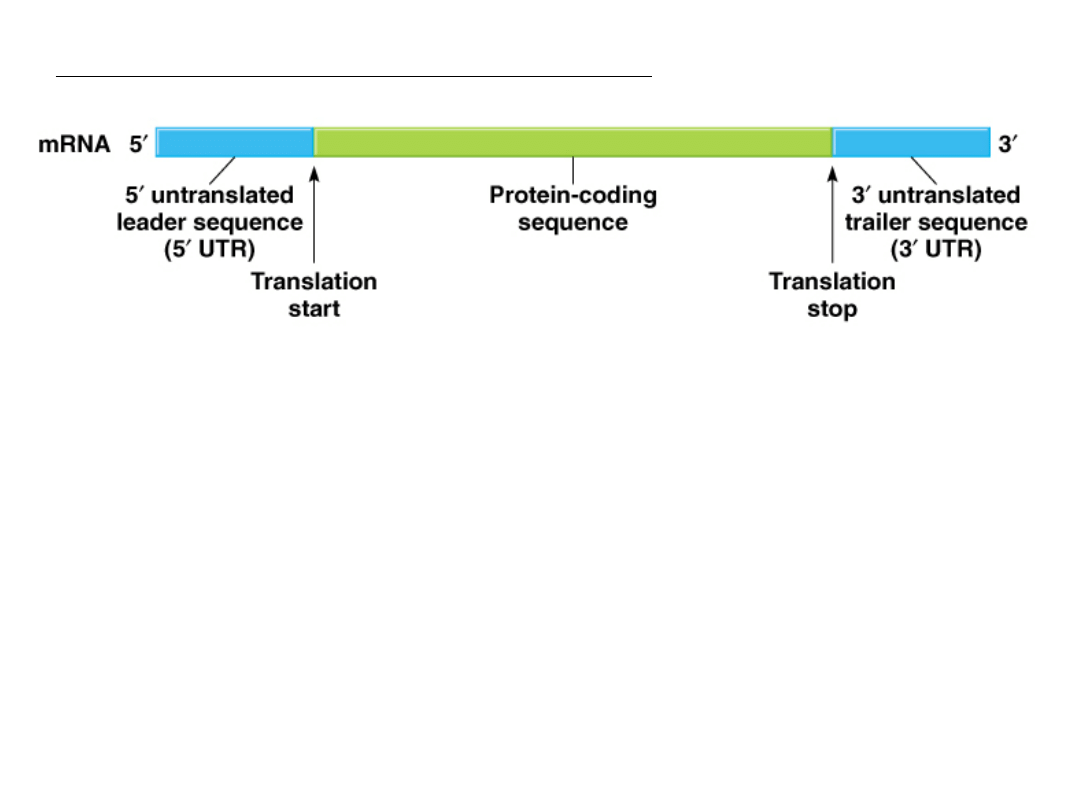
Production of the mRNA molecule (Fig. 5.8)
Three main parts:
1.
5’ untranslated region (5’ UTR) or leader sequence
2.
Coding sequence, specifies amino acids to be translated
3.
3’ untranslated region ( 3’ UTR) or trailer sequence
may contain information that signals the stability of the
particular mRNA
mRNA differences between prokaryotes and eukaryotes:
Prokaryotes
1.
mRNA transcript is mature, and used directly for translation
without modification.
2.
Since prokaryotes lack a nucleus, mRNA also is translated on
ribosomes before it is transcribed completely (i.e., transcription
and translation are coupled).
3.
Prokaryote mRNAs are polycistronic, they contain amino acid
coding information for more than one gene.
Eukaryotes
1.
mRNA transcript is not mature (pre-mRNA); must be processed.
2.
Transcription and translation are not coupled (mRNA must first be
exported to the cytoplasm before translation occurs).
3.
Eukaryote mRNAs are monocistronic, they contain amino acid
sequences for just one gene.
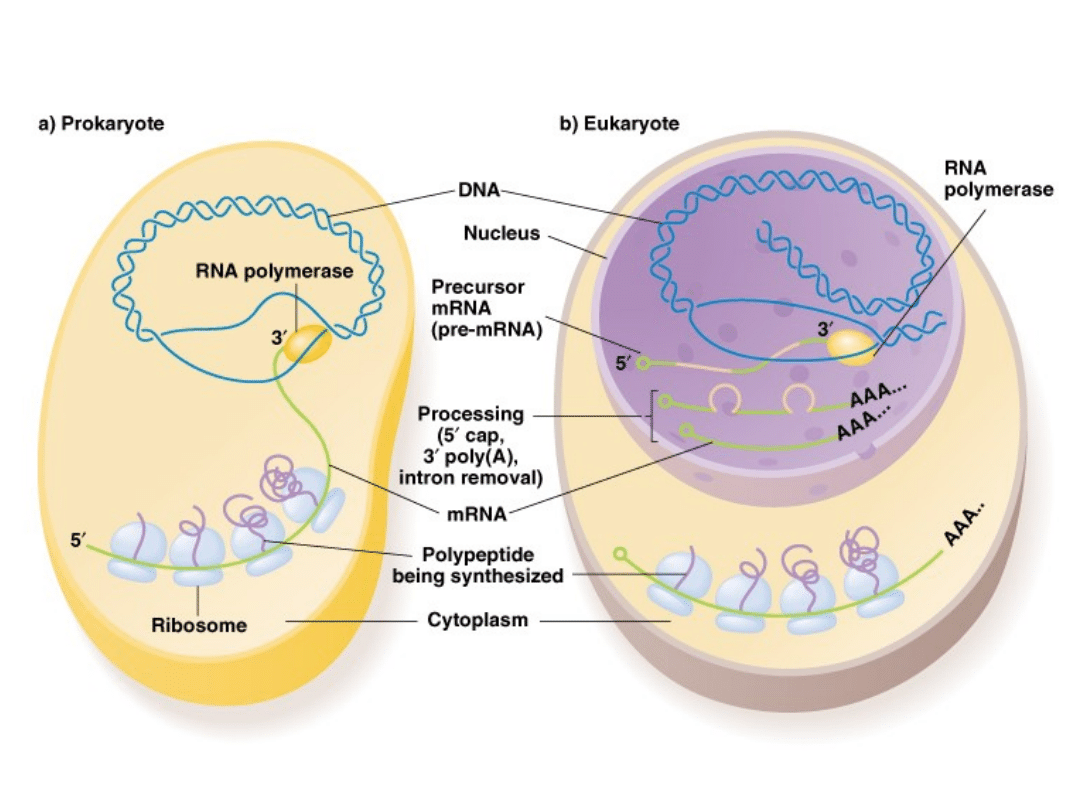
Fig. 5.9. Prokaryotes and Eukaryotes

Production of mature mRNA in eukaryotes:
1.
5’ cap
After 20-30 nucleotides have been synthesized, the 5’-end of
the mRNA is capped 5’ to 5’ with a guanine nucleotide (See
Fig. 5.10).
Results in the addition of two methyl (CH
3
) groups.
Essential for the ribosome to bind to the 5’ end of the mRNA.
2.
Poly (A) tail
50-250 adenine nucleotides are added to 3’ end of mRNA.
Complex enzymatic reaction.
1.
Stabilizes the mRNA, and plays an important role in
transcription termination.
•
Introns (non-coding sequences between exons) are removed and
exons (amino acid coding sequences) are spliced.

Animation RNA processing
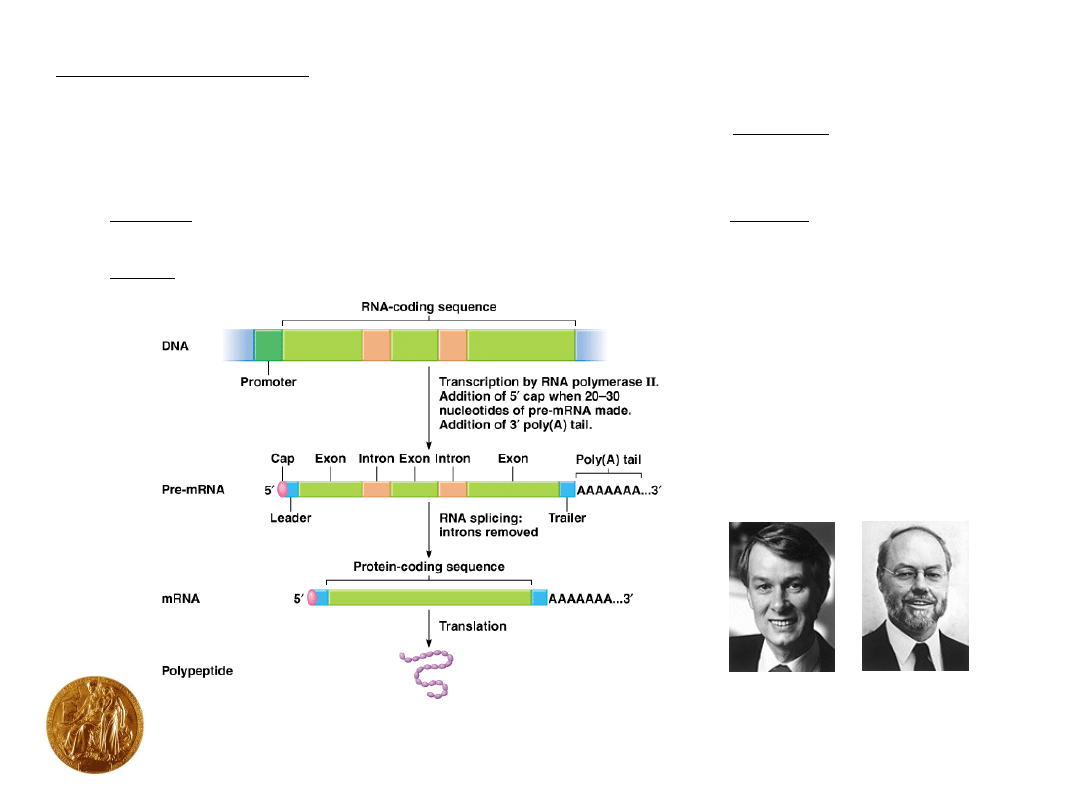
Introns and exons:
Eukaryote pre-mRNAs often have intervening introns that must be
removed during RNA processing (as do some viruses).
intron = non-coding DNA sequences between exons in a gene.
exon = expressed DNA sequences in a gene, code for amino acids.
1993: Richard Roberts (New England Biolabs) & Phillip Sharp (MIT)
Fig. 5.12
mRNA splicing of exons and removal of introns:
1.
Introns typically begin with a 5’-GU and end with AG-3’.
2.
Cleavage occurs first at the 5’ end of intron 1 (between 2 exons).
3.
The now free G joins with an A at a specific branch point sequence
in the middle of the intron, using a 2’ to 5’ phosphodiester bond.
Intron forms a lariat-shaped structure.
4.
Lariat is excised, and the exons are joined to form a spliced mRNA.
5.
Splicing is mediated by splicosomes, complexes of small nuclear
RNAs (snRNAs) and proteins, that cleave the intron at the 3’ end
and join the exons.
6.
Introns are degraded by the cell.
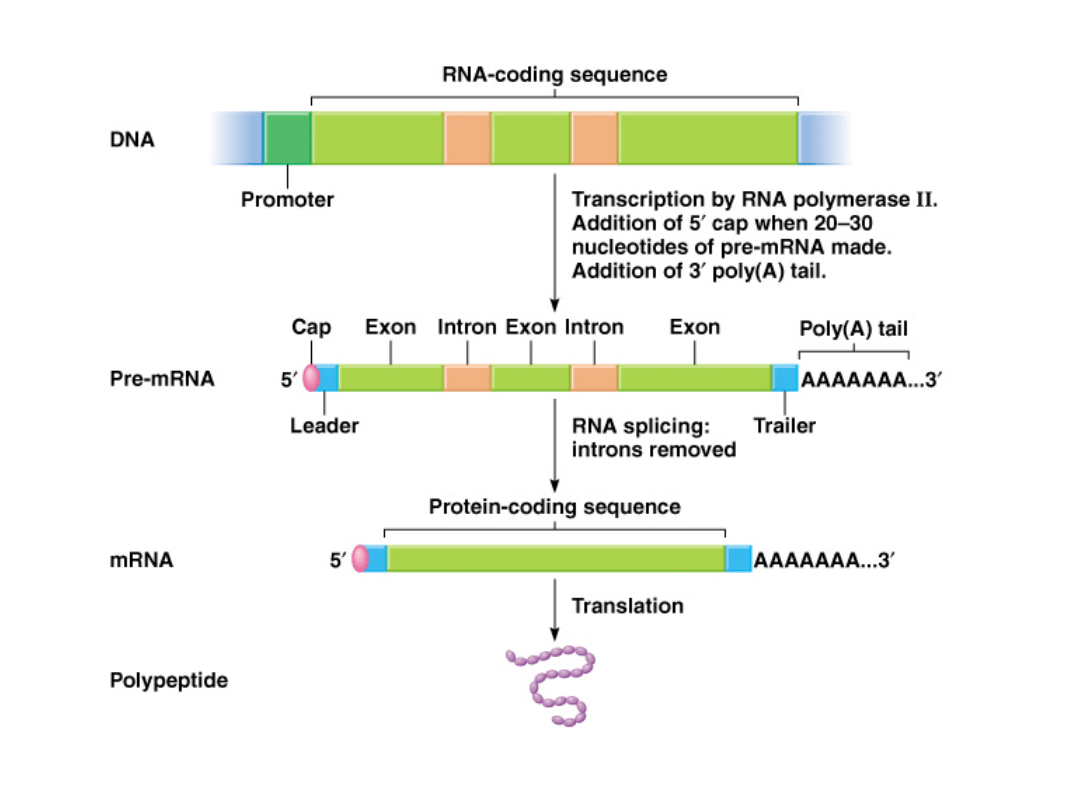
Fig. 5.12
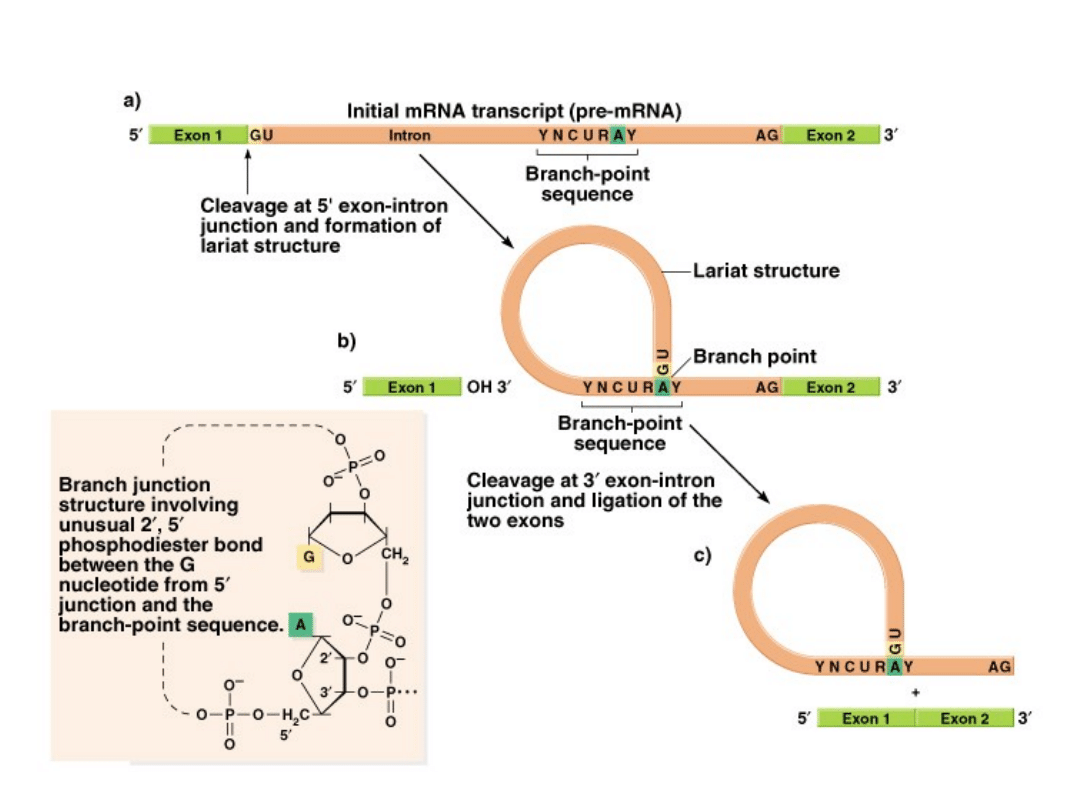
Fig. 5.13

Animation Intron Removal

Post-transcriptional modification, mRNA editing:
1.
Adds or deletes nucleotides from a pre-RNA, or chemically alters
the bases, so the mRNA bases do not match the DNA sequence.
2.
Can results in the substitution, addition, or deletion of amino acids
(relative to the DNA template).
3.
Generally cell or tissue specific.
4.
Examples occur in protozoa, slime molds, plant organelles, and
mammals.

Genes that do not code proteins also are transcribed:
1.
rRNA, ribosomal RNA
Catalyze protein synthesis by facilitating the binding of tRNA
(and their amino acids) to mRNA.
2.
tRNA, transfer RNA
Transport amino acids to mRNA for translation.
3.
snRNA, small nuclear RNA
Combine with proteins to form complexes used in RNA
processing (splicosomes used for intron removal).

1. Synthesis of ribosomal RNA and ribosomes:
1.
Cells contain thousands of ribosomes.
2.
Consist of two subunits (large and small) in prokaryotes and
eukaryotes, in combination with ribosomal proteins.
3.
E. coli 70S model:
50S subunit = 23S (2,904 nt) + 5S (120 nt) + 34 proteins
30S subunit = 16S (1,542 nt) + 20 proteins
4.
Mammalian 80S model:
60S subunit = 28S (4,700 nt) +5.8S (156 nt) + 5S (120 nt) +
50 proteins
40S subunit = 18S (1,900 nt) + 35 proteins
5.
DNA regions that code for rRNA are called ribosomal DNA (rDNA).
6.
Eukaryotes have many copies of rRNA genes tandemly repeated.
1. Synthesis of ribosomal RNA and ribosomes (continued):
7.
Transcription occurs by the same mechanism as protein-coding
genes, but generally using RNA polymerase I.
8.
rRNA synthesis requires its own array transcription factors (TFs)
9.
Coding sequences for RNA subunits within rDNA genes contain
internal (ITS), external (ETS), and nontranscribed spacers (NTS).
10. ITS units separate the RNA subunits through the pre-rRNA stage,
whereupon ITS & ETS are cleaved out and rRNAs are assembled.
11. Subunits of mature ribosomes are bonded together by H-bonds.
12. Finally, transported to the cytoplasm to initiate protein
synthesis.
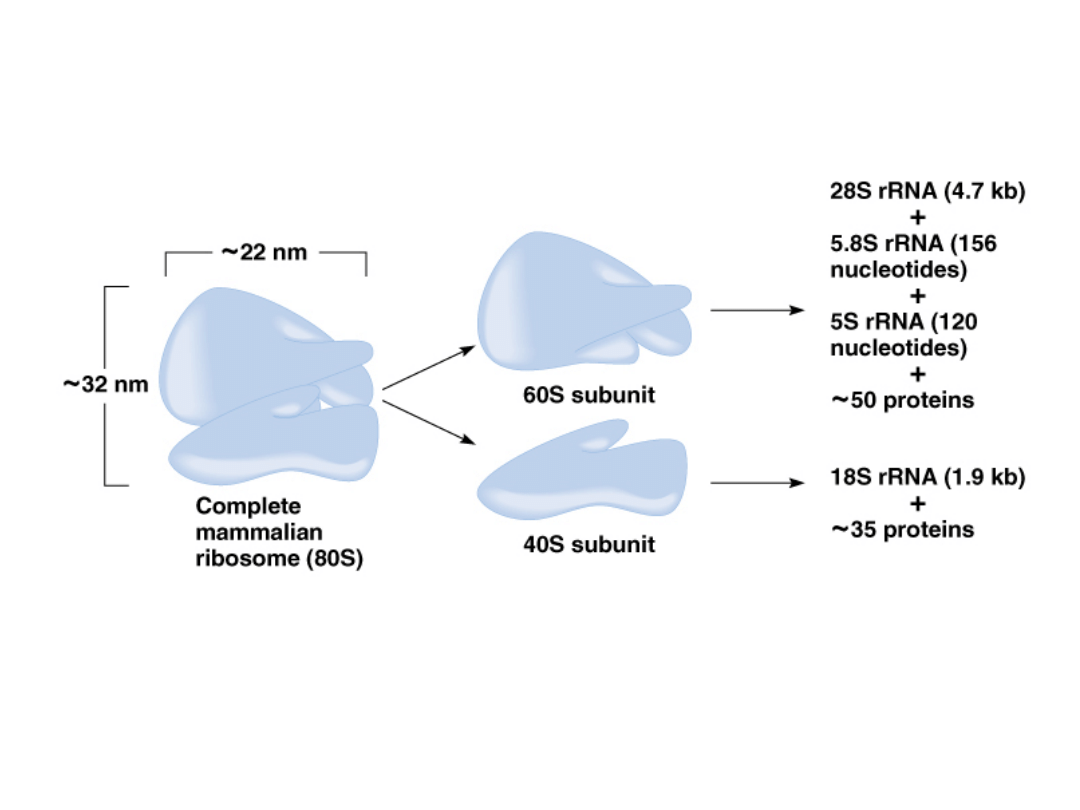
Fig. 6.13, Mammalian example of 80S rRNA
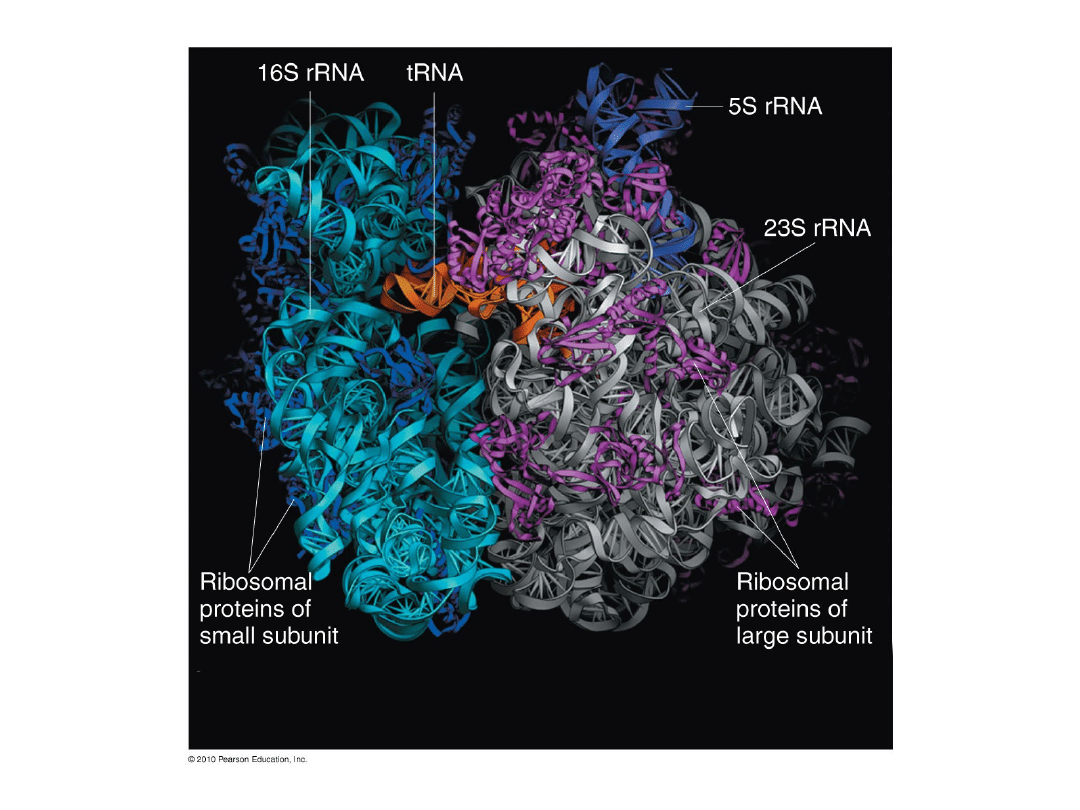
Fig. 6.12
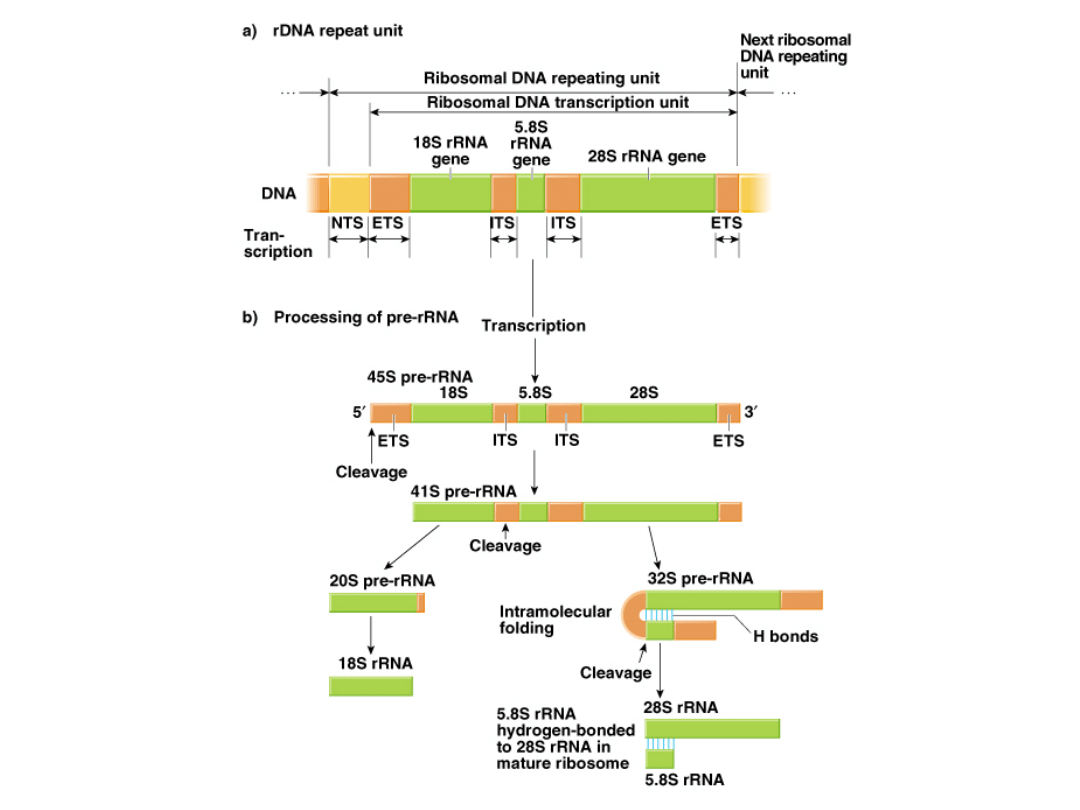
Fig. 5.18
2nd
edition

2. Synthesis of tRNA:
1.
tRNA genes also occur in repeated copies throughout the
genome, and may contain introns.
2.
Each tRNA (75-90 nt in length) has a different sequence that
binds a different amino acid.
3.
Many tRNAs undergo extensive post-transcription modification,
especially those in the mitochondria and chloroplast.
4.
tRNAs form clover-leaf structures, with complementary base-
pairing between regions to form four stems and loops.
5.
Loop #2 contains the anti-codon, which recognizes
mRNA codons during translation.
6.
Same general mechanism using RNA polymerase III, promoters,
unique TFs, plus posttranscriptional modification from pre-tRNA.
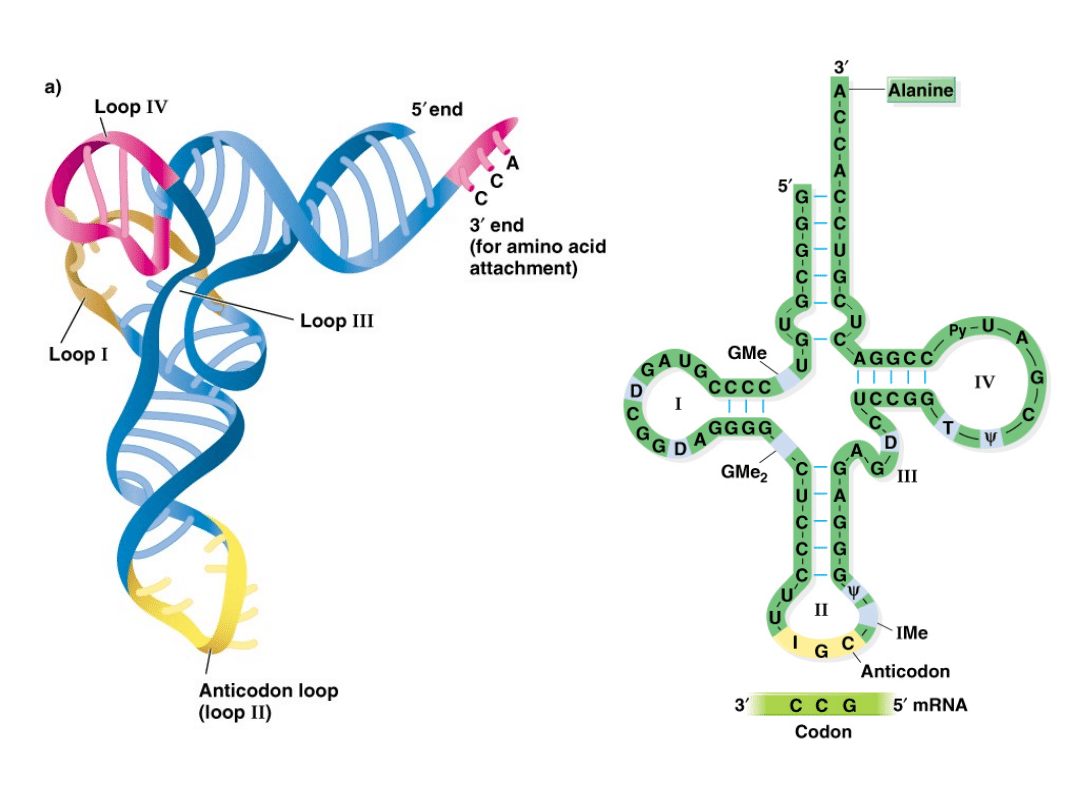
Figure 6.9

3. Synthesis of snRNA (small nuclear RNA):
•
Form complexes with proteins used in eukaryotic RNA processing,
such as splicing of mRNA after introns are removed.
•
Transcribed using RNA polymerase II or III.
•
Associated with small nuclear ribonucleoproteins (snRNPs).
•
Also function in regulation of transcription factors and
maintenance of telomeres.
Document Outline
- PowerPoint Presentation
- Slide 2
- Slide 3
- Slide 4
- Slide 5
- Slide 6
- Slide 7
- Slide 8
- Slide 9
- Slide 10
- Slide 11
- Slide 12
- Slide 13
- Slide 14
- Slide 15
- Slide 16
- Slide 17
- Slide 18
- Slide 19
- Slide 20
- Slide 21
- Slide 22
- Slide 23
- Slide 24
- Slide 25
- Slide 26
- Slide 27
- Slide 28
- Slide 29
- Slide 30
- Slide 31
- Slide 32
- Slide 33
- Slide 34
- Slide 35
- Slide 36
- Slide 37
- Slide 38
Wyszukiwarka
Podobne podstrony:
Feynman Lectures on Physics Volume 1 Chapter 05
Dictionary Chapter 05
Intro to ABAP Chapter 05
chapter 05 conversions
English Skills with Readings 5e Chapter 05
Fundamentals of College Physics Chapter 05
Essential College Experience with Readings Chapter 05
durand word files Chapter 5 05753 05 ch5 p172 207 Caption
durand word files Chapter 5 05753 05 ch5 p172 207
Fundamentals of Anatomy and Physiology 05 Chapter
Figures for chapter 5
podrecznik 2 18 03 05
regul praw stan wyjątk 05
Figures for chapter 12
więcej podobnych podstron