
Genetic Selection Tools in
the functional Genomics

Outline
Background
–
Genetic Evaluations
–
Quantitative Genetics
–
Genomics
Integrating Genetics and Genomics
Case Study: DGAT1
Tangent: Animal Identification
Crystal Ball
Conclusions

Background
Bovine Functional Genomics Laboratory (BFGL)
–
Structural and functional genomics of cattle
–
Emphasis on health and productivity
–
Bioinformatics (storage and use of genomic data)
Animal Improvement Programs Laboratory (AIPL)
–
“Traditional” genetic improvement of dairy cattle
–
Increasing emphasis on animal health and reproduction

Traditional Selection
Programs
Estimate genetic merit for animals in a
population
Select superior animals as parents of
future generations

Genetic Evaluation System
Traditional selection has been very effective
for many economically important traits
Example: Milk yield
–
Moderately heritable
–
~30 million animals evaluated 4x/yr
–
Uses ~70 million lactation records
–
Includes ~300 million test-day records
–
Genetic improvement is near theoretical
expectation
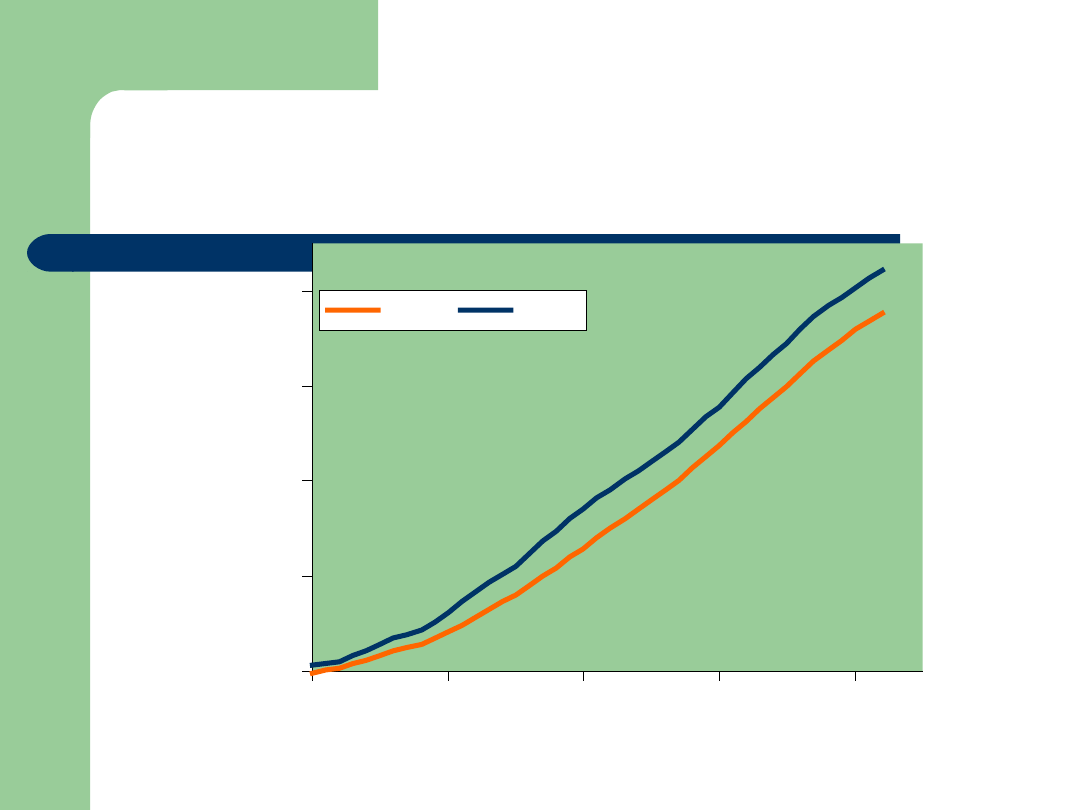
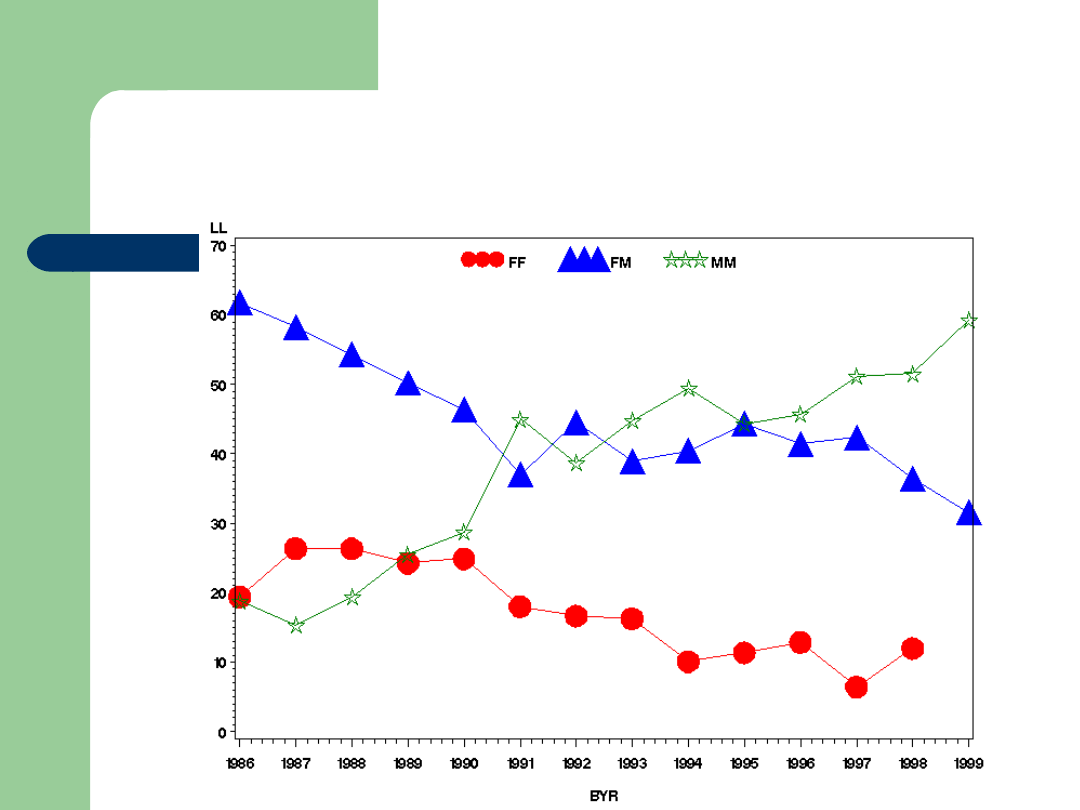
Dairy Cattle Genetics Success
-6000
-4000
-2000
0
2000
1960
1970
1980
1990
2000
Year of Birth
B
V
M
il
k
Cows
Bulls
Dairy Cattle Genetics
Industry Cooperation
Producer
DHIA/DRPC
PDCA
NAAB
AIPL
Producer
DHIA/DRPC
PDCA
NAAB
AIPL

Genomics - Introduction
Traditional dairy cattle breeding has assumed
that an infinite number of genes each with
very small effect control most traits of
interest
Logical to expect some “major” genes with
large effect; these genes are usually called
quantitative trait loci (
QTL
)
The QTL locations are unknown!
Genetic markers can provide information
about QTL
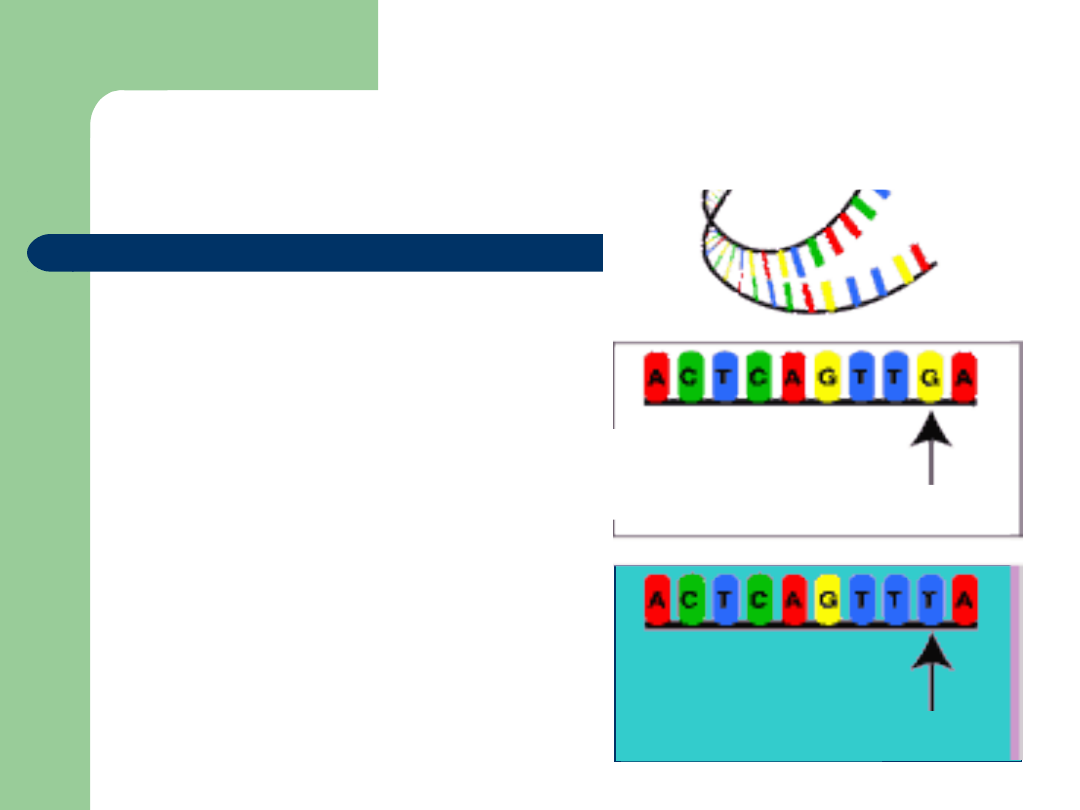
Genetic Markers
Allow inheritance of a
region of the genome to be
followed across generations
Single nucleotide
polymorphisms (
SNiP
) are
the markers of the future!
Need lots!
–
3 million in the genome
–
10,000 initial goal
Polymorphis
m
“poly”
= many
“morph”
=
form
General
population
94%
6%
Single nucleotide
polymorphism
(SNP)

Application of Genetic
Markers
1.
Identify genetic markers or polymorphisms in
genes that are associated with changes in
genetic merit
2.
Use marker assisted selection (
MAS
) or gene
assisted selection (
GAS
) to make selection
decisions before phenotypes are available
3.
Adjust genetic merit for markers or genes in
the genetic evaluation system
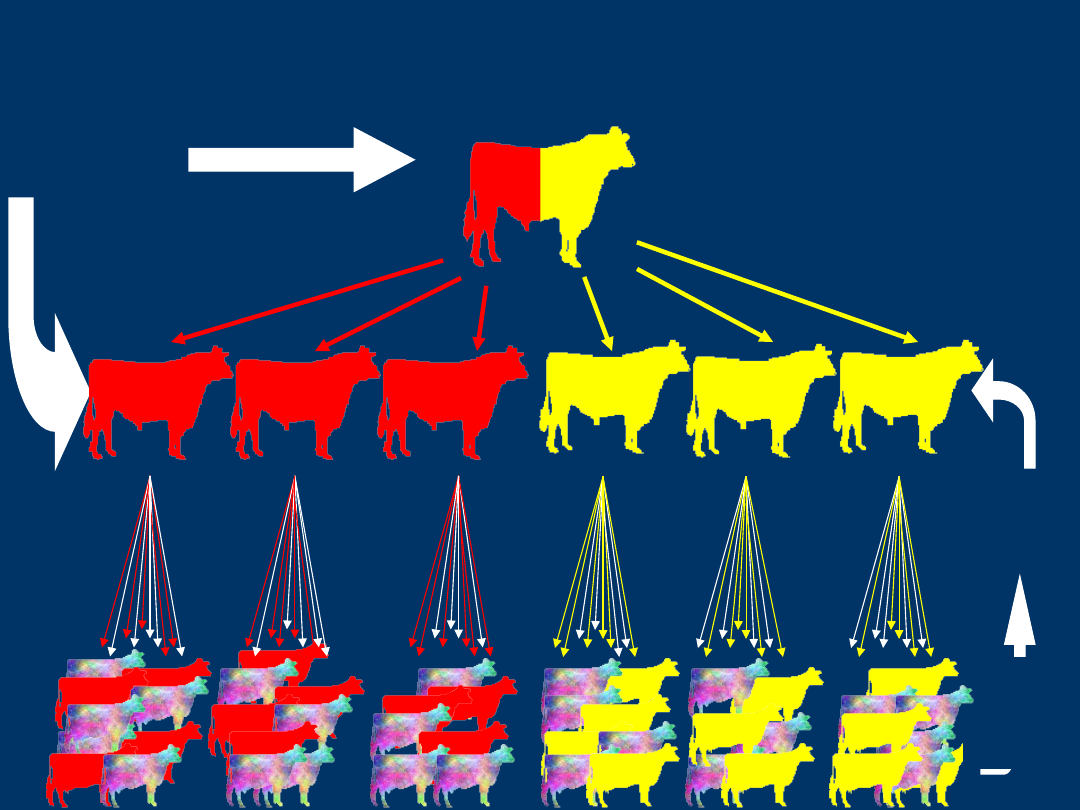
QTL Identification
Geneti
c
Merit
DNA
Dat
a
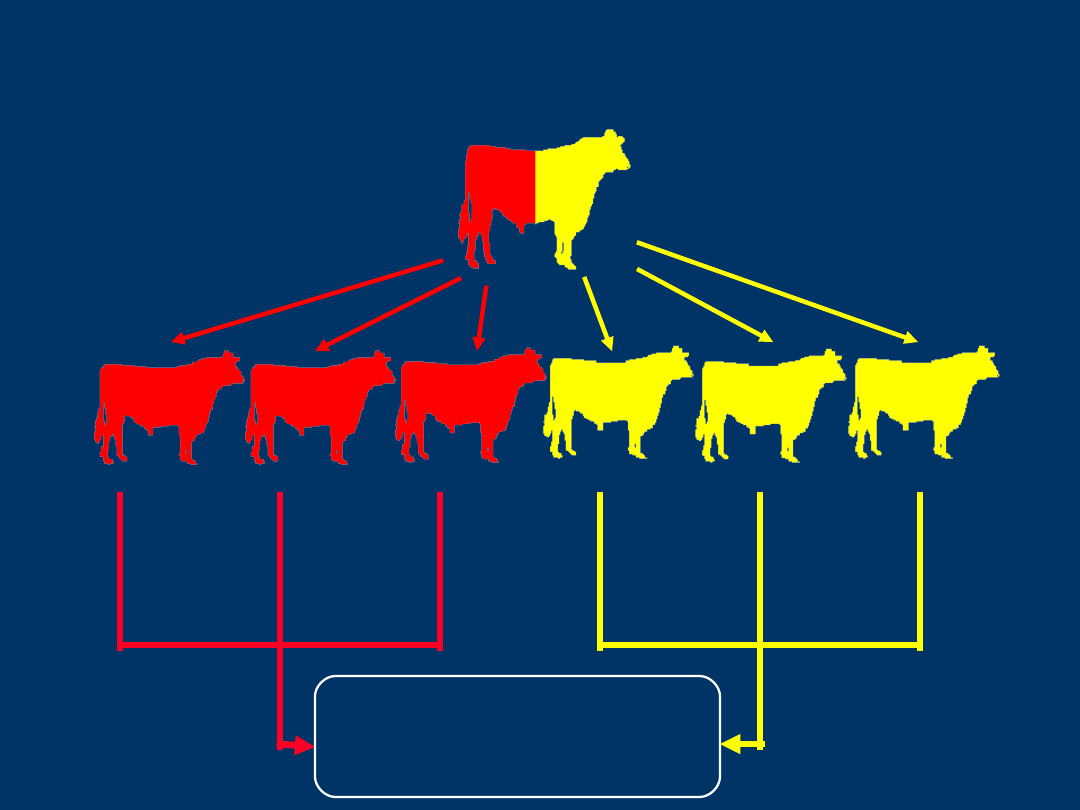
Compare Genetic
Merit
QTL Identification and
Marker Assisted Selection
3.5
1.7
-0.1
-2.5
-6.2
0.7
Gene Assisted
Selection

Marker or Gene Assisted
Selection
Largest benefits are for traits that:
–
have low heritability, i.e., traits where genetics
contribute a small fraction of observed variation (e.g.,
disease resistance and fertility)
–
are difficult or expensive to measure (e.g., parasite
resistance )
–
cannot be measured selection decision needs to be
made (e.g., milk yield and carcass characteristics)
Evolution in traditional selection program by
improving estimation of genetic merit

Example: DGAT1
DGAT1: diacylglycerol
acyltransferase
–
Enzyme involved in fat sythesis
–
Identified using
Genetic marker data
Model organism (mouse) gene function
information
Cattle sequence verified candidate gene

DGAT1
Two forms of the gene in cattle
–
M = high milk (low fat) form of gene
–
F = high fat (low milk) form gene
BFGL scientists decided to characterize
the gene in North American population
–
Over 3300 animals genotyped for DGAT1 SNP
–
Approximately 2900 genotypes verified and
used in these analyses

DGAT1 – Average Differences
in Daughters of Bulls
Trait
MM-
FF
Trait
MM-
FF
Milk lbs
361
Fat%
0.13
Fat lbs
16.5
Protein
%
0.02
Protein
lbs
5.0
NM$
$24
SCS
0.05
CM$
$35
PL
0.07
FM$
$4
DPR
0.21
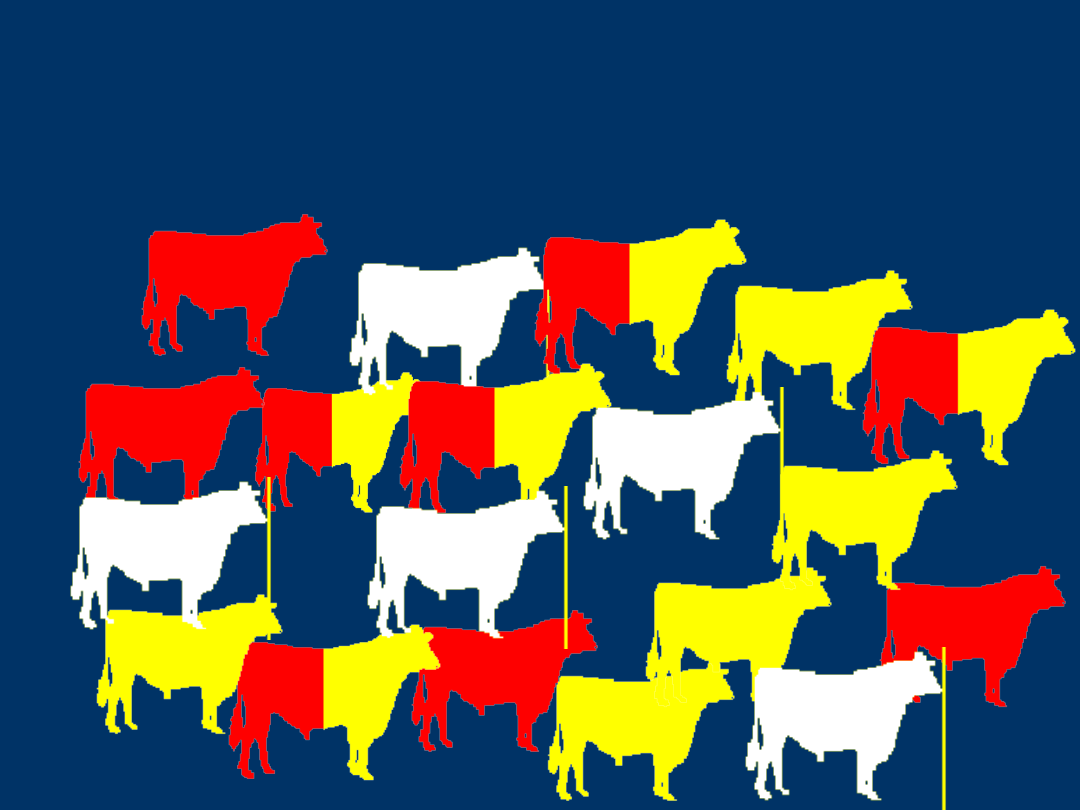
DGAT1 Genotypic
Frequencies

Integrating Genomics
Results
Genes will likely account for a
fraction of the total genetic
variation
Cannot select solely on gene
tests!!
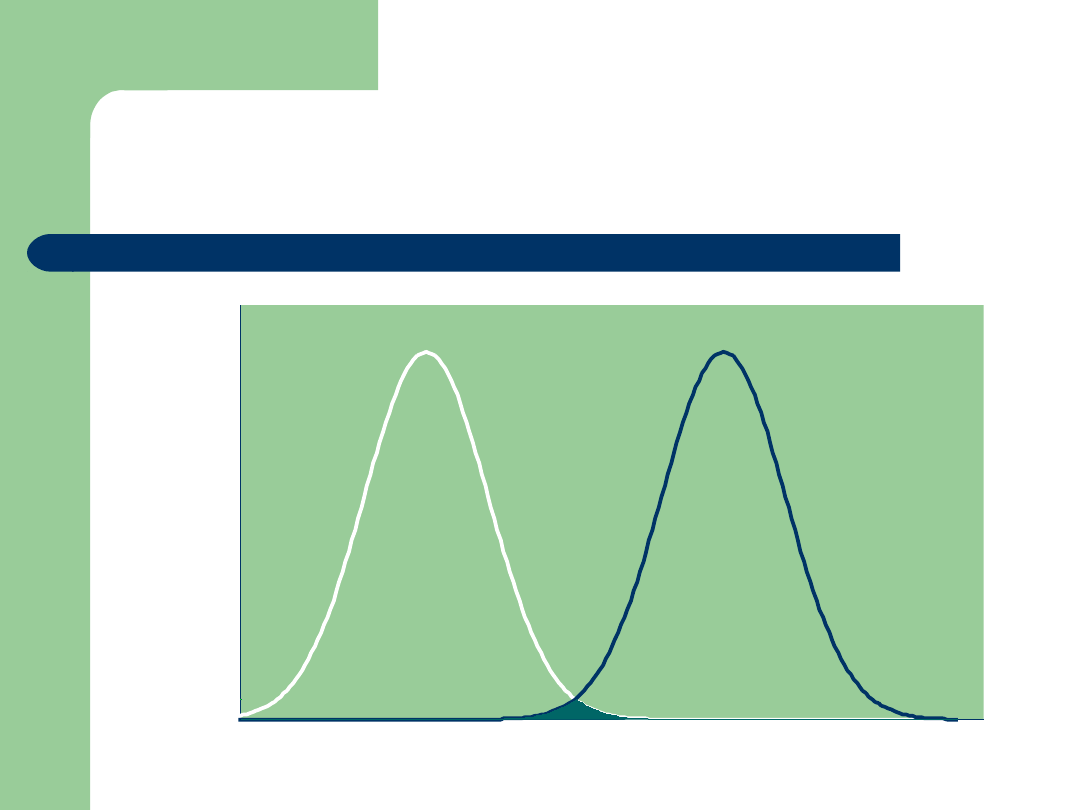
Integrating Genomic Data:
An Ideal Situation!
Bull PTA
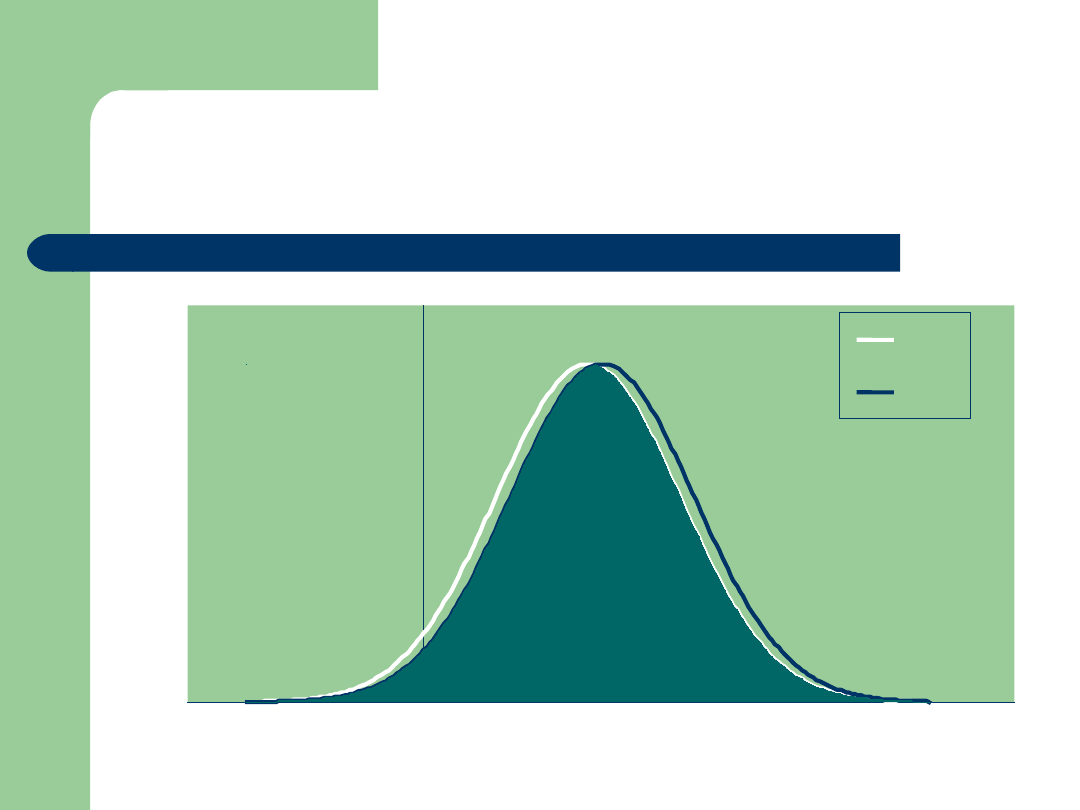
Integrating Genomic Data:
The DGAT1 NM$ Situation!
Bull PTA NM$
MM
FF
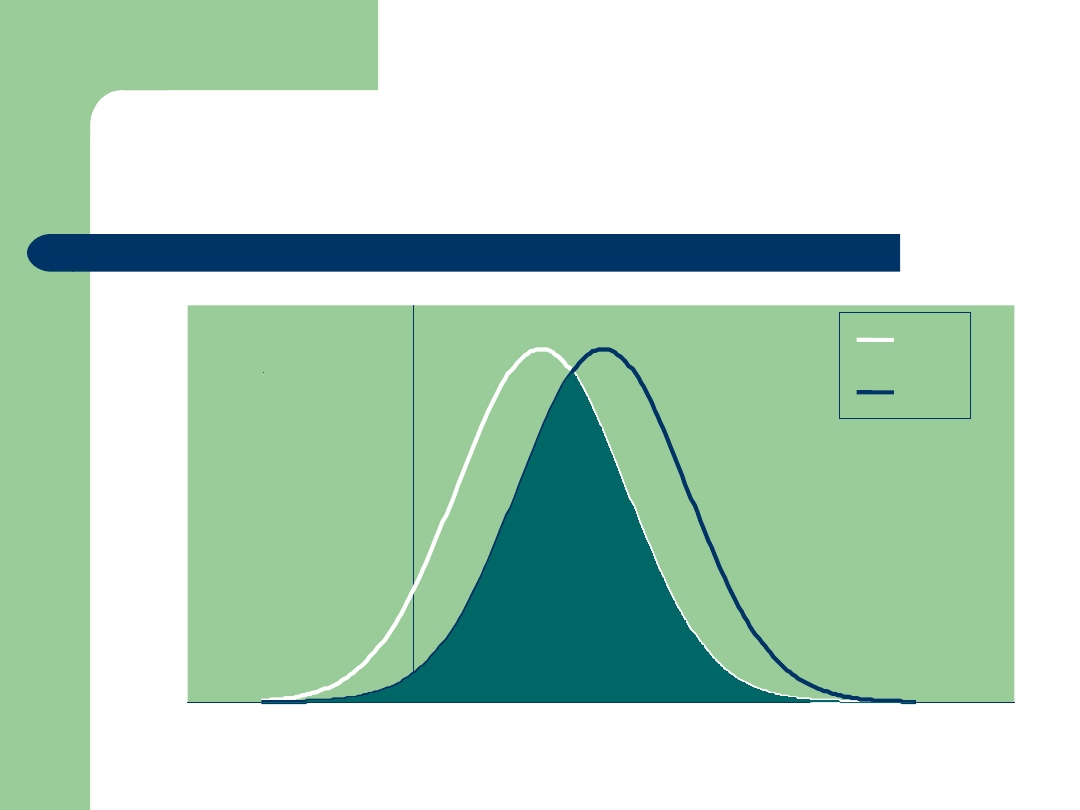
Integrating Genomic Data:
The DGAT1 Fat Situation!
Bull PTA Fat
MM
FF

Integrating Genomics
Results
Combine information
–
Ideally would incorporate genomic data into
genetic evaluation system
Adjust PTA??
–
Don’t adjust well proven animals (it’s in
there!!)
–
Adjust parent average for flush mates
–
Progeny have identical parent averages
–
Adjusting other PTA is non-trivial!
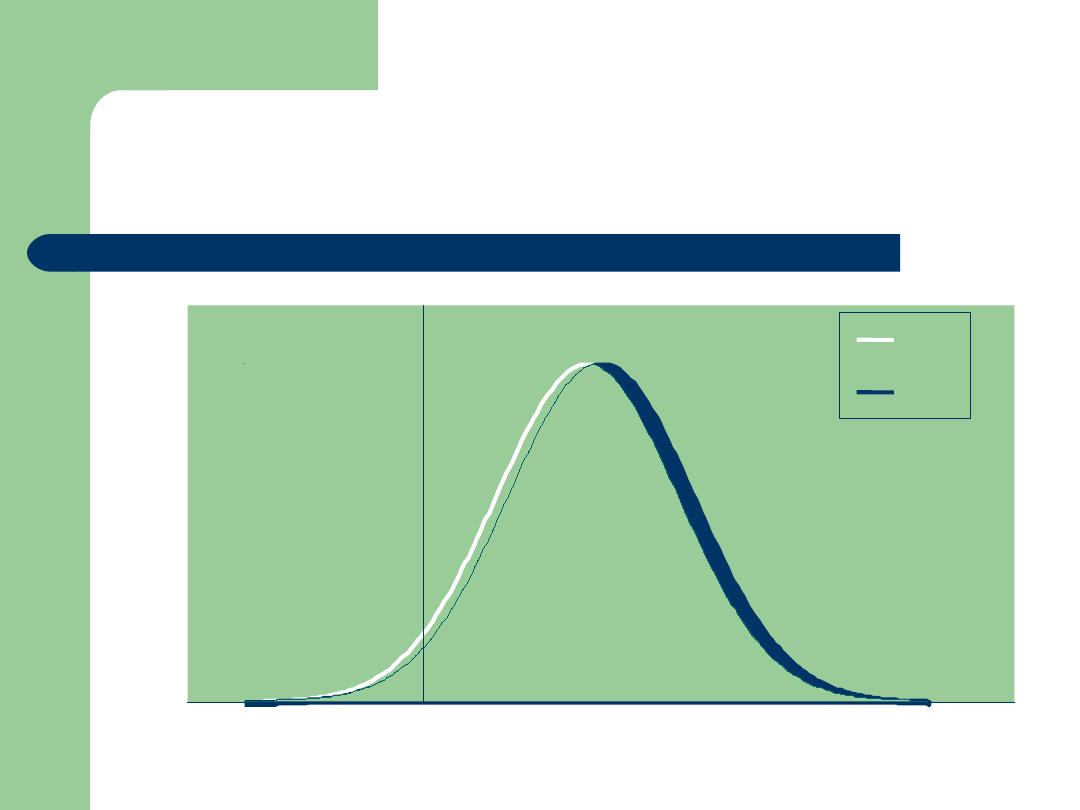
Integrating Genomic Data:
Another view of DGAT1 NM$!
Bull PTA NM$
MM
FF

And it Really Works!
Recent German study evaluated impact on
adjusting historic parent averages (
PA
) for DGAT1
and evaluated impact of predictability of future
evaluations
Correlations of original PA with eventual PTA for milk
were 45%
Correlations of adjusted PA with eventual PTA for
milk were 55% (
10% gain
)
Incorporation of genomic data will result in
increased stability of evaluations

Genetic Evaluations -
Limitations
Slow!
–
Progeny testing for production traits take 3 to 4
years from insemination
–
A bull will be at least 5 years old before his
first evaluation is available
Expensive!
–
Progeny testing costs $25,000 per bull
–
Only 1 in 8 to 10 bulls graduate from progeny
test
–
At least $200,000 invested in each active bull!!

Genetic Evaluations:
Genomics Enhancements
Faster
–
Use of gene and marker tests allow
preliminary selection decisions beyond
parent average before performance and
progeny test data are available
Cheaper
–
Improved selection decisions should result
in higher graduation rates or enhanced
genetic improvement

How do we get there
Increase number of genetic markers
Continue QTL discovery for MAS/GAS
Better characterize the genome
–
Compare genome to well characterized
human and mouse genome

Bovine Genome Sequence

Bovine Genome Sequence
Inbred Hereford is primary animal being
sequenced
Genome size is similar to humans
Sequencing about half completed
First assembly released yesterday!!
–
2.3 of 2.8 billion base pairs
–
84% coverage
L1 Dominette 01449

Bovine Genome Sequence
Six breeds selected for
low level sequencing
Holstein and Jersey cows
represent dairy breeds
Useful for SNP marker
development
Expect 3 million SNPs in
the genome
Preliminary goal is to
characterize 10,000
Wa-Del RC Blckstr Martha-ET
Mason Berretta Jenetta

Genomic Tools for
Parentage Verification
Low-cost high-throughput SNP marker tests would
facilitate parentage verification and traceability
$10 to $20 per sample seems to be a common break
point
Progeny test herds would likely be early adopters
–
Support from studs?
Results in increased stability on first proofs?
–
Nearly impossible to make mistake on parentage
–
Punished on second crop proofs?
With widespread implementation
–
Increase effective heritability
–
Decrease evaluation variability
–
Enhanced genetic improvement

Crystal Ball (Wishful
Thinking?)
Large number of validated genetic tests
available
Large amounts of marker and gene
data publicly available
Genomic data incorporated into genetic
evaluation
Management decisions facilitated by
genomics data

Considerations in Genomic
Tests
How big is the effect?
–
Traits of interest, economic index (NM$, TPI, PTI)
–
How many genetic standard deviation units?
Has this been validated by a sufficiently large
independent study?
What correlated response is expected & observed?
What are allele frequencies?
What is the value of this test?
–
not simple to answer

Conclusions
Genomics is enhancing genetic improvement
DGAT1 has large impacts on milk, fat,
protein, SCS
Genetic tests need to be weighted
appropriately for optimal selection decisions
Genomic tools will be extremely powerful for
parentage verification and traceability
–
Could impact genetic evaluations
Document Outline
- Genetic Selection Tools in the functional Genomics
- Outline
- Background
- Traditional Selection Programs
- Genetic Evaluation System
- Dairy Cattle Genetics Success
- Dairy Cattle Genetics Industry Cooperation
- Genomics - Introduction
- Genetic Markers
- Application of Genetic Markers
- Slide 11
- QTL Identification and Marker Assisted Selection
- Gene Assisted Selection
- Marker or Gene Assisted Selection
- Example: DGAT1
- DGAT1
- DGAT1 – Average Differences in Daughters of Bulls
- DGAT1 Genotypic Frequencies
- Integrating Genomics Results
- Integrating Genomic Data: An Ideal Situation!
- Integrating Genomic Data: The DGAT1 NM$ Situation!
- Integrating Genomic Data: The DGAT1 Fat Situation!
- Slide 23
- Integrating Genomic Data: Another view of DGAT1 NM$!
- And it Really Works!
- Genetic Evaluations - Limitations
- Genetic Evaluations: Genomics Enhancements
- How do we get there
- Bovine Genome Sequence
- Slide 30
- Slide 31
- Genomic Tools for Parentage Verification
- Crystal Ball (Wishful Thinking?)
- Considerations in Genomic Tests
- Conclusions
Wyszukiwarka
Podobne podstrony:
Nonlinear Control of a Conrinuously Variable Transmission (CVT) for Hybrid Vehicle Powertrains
MITSUBISHI EXPO EXPO LRV 1992
MITSUBISHI EXPO 1995
MITSUBISHI EXPO EXPO LRV 1993
AMC CVT 2100 service manual
CVT,CVTO
Caliber CVT Operation
MITSUBISHI EXPO EXPO LRV 1994
Ciekawostki targów innowacji w Amber Expo
Światowa Wystawa EXPO 2005 Japonia, 2005
więcej podobnych podstron