browser
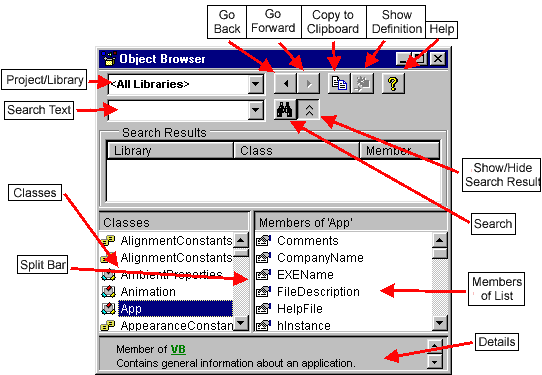
Splil Bar
Classes
AlignmentConstants 1 AlignmentConstants—1 ~~ I1*'! ml i niri i^rtinr p @ Animation
isP AppearanceConstariZJ
Members of App' & Comments CompanyName ® EKEName d? FileDescription ® HelpFile hlnstance
Member of VB
Contains generał information about an application.
|
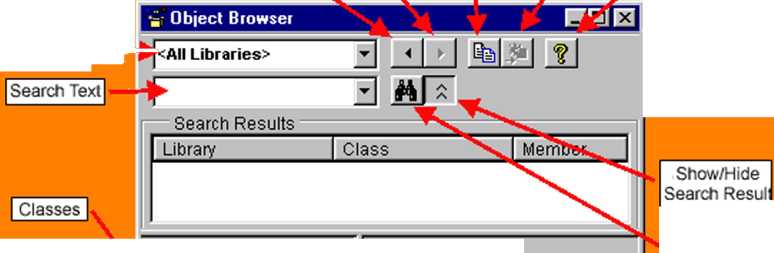
s| Search | ||
|
Members of List | ||
|
Details | ||
Wyszukiwarka
Podobne podstrony:
PTC027 .i i i n - II! I ;i I I i dlii I I I ! -I—1—r ML. i ! I cl A t. ^ %e^ f kIW Ci-bi I !u 1 J
Instrukcja FOTO bmp i* Add morę languages r . I1. Ml . : Hełp Bcustormze Preference Translate thłs
•(«*) • *■*1^ I1 i I1• mł) I » •<■*) > Ml1) ^
•(«*) • *■*1^ I1 i I1• mł) I » •<■*) > Ml1) ^
f fig13 Add Member Variable Member variable name:
restrictionmap BioEdit Sequence Alignment Editor - [Restriction Map: Humań coronavirus NL63 isoiate
29.10.2015 Bioinformatyka, edycja 2015/2016, laboratorium 4Global Alignment vs. Local
Paragraphfe, W Background Indents & Spacinj
A J2EE based server for Muon Spectrometer Alignment monitoring in the ATLAS detector Journal of
Figurę 1. Sagitta definition and Figurę 2. ATLAS muon spectrometer optical alignment system. optical
5) Oracle Database for the alignment monitoring tables, optical sensors data and conditions data, ma
Figurę 5. Alignment corrections tables structure. Figurę 6. AłignGUI example functionalities. 6.
więcej podobnych podstron