3416400799
NF-kB and radioscnsi(ivity • C. DlDELOT et al. 1355
mediated transcriptional activation and provides a fcedback mechanism.
Because NF-kB was activated in numerous cancers, it might confer intrinsic radioresistance. In this study, wc investigated the effect of modulation of the NF-kB DNA-binding activity on the intrinsic radiosensitivity of head-and-neck celi Iines. We recently reported that the cellular radiosensitivity of head-and-neck carcinoma celi lines was related to baseline apoptosis (16). In the present study, we investigated whether modulation of constitutive NF-kB ac-tivity by dexamethasone (DEX) and TNFa could modify the intrinsic radiosensitivity and the basal apoptosis of head-and-neck carcinoma celi lines.
MATERIALS ANI) METHODS
Celi culture materials were purchased from Costar (Dut-scher, Brumath, France), and culture media and additives were obtained from Life Technologies (Gibco BRL, Cergy-Pontoise, France), except fetal calf serum, which was obtained from Costar (Dutscher, Brumath, France). Recombi-nant hurnan TNFa was provided by RD Systems (Abingdon, UK). Antibodies against P50 and P65 were purchased from Santa Cruz (Tebu, Le Perray-en-YveIines, France). Ali other Chemicals were purchased from Sigma (St. Quentin Fallavier, France).
Celi culture and treatment
KB head-and-neck carcinoma celi linę was kindly pro-✓ided by A. Hanauske (Munich University, Germany) as lart of the EORTC Preclinical Therapeutic Models Group 3xchange program. The KB3 subline was derived from KB ;ells, as previously described (16). Both celi lines were »rown in a 37°C, 5% C02 atmosphere in 75-cm2 plastic issue culture flasks in RPMI 1640 medium supplemented vith 10% fetal calf serum, penicillin (100 i.u7mL), and .treptomycin (100 jitg/mL).
Modulation of NF-kB DNA-binding activity
KB and KB3 celi lines were secded at 2.104 cells/cm2. ?our days later, the cells were exposed to 1 yxM DEX for !4 h or 2.5 ng/mL TNFa for 10 min. After this treatment, he cells were culturcd in drug-free medium for 14 h.
dectrophoretic mobility shift and supershift assays
Nuclear extracts from KB and KB3 cells exposed to 'NFa or DEX were prepared as described previously 17), with the following modifications. The nuclei were esuspendcd in 20 mM HEPES, pH 7.9, containing 450 iM NaCl, 0.2 mM EDTA, 0.15 mM MgCl2, 25% (v/v) lycerol, 0.5 mM dithiothreitol, and 0.5 mM phenylmeth-Isulfonyl sulforidc, then homogeneized in Dounced Fisher Bioblock Scientiłic, Illkirch, France) and incu-ated for 30 min at 4°C after addition of 0.25% (v/v) Jonidet P-40. Nuclear extracts were eleared by centrif-gation for 30 min at 4°C at 17,000 g, and the resulting upernatants were supplemented with KC1 (0.1 M) and
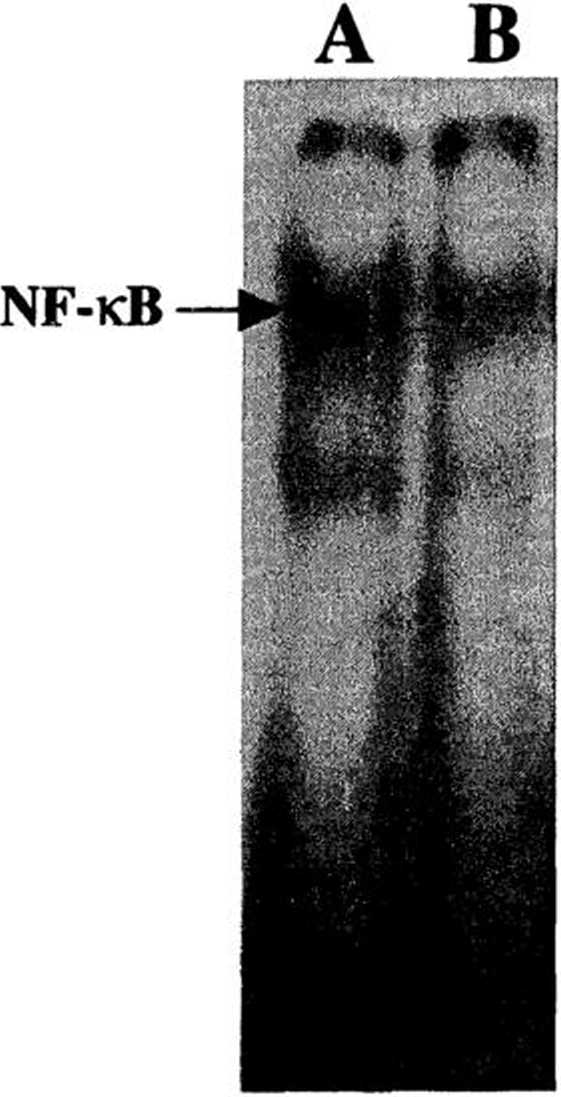
Fig. 1. Constitutive DNA-binding activity of NF-kB in KB and KB3 cells. Nuclear extracts (5 pg) from (A) KB and (B) KB3 were ineubated with radiolabclcd NF-kB binding site. The DNA/NF-kB complcx was analyzed in nativc polyacrylamide gcls. This picture was representative of threc independent experiments.
immcdiately frozen in liquid nitrogen and stored at -20°C until used for electrophoretic mobility shift assay and supershift assay.
Nuclear protein concentration was determined according to Lowry et al. (18) using bovine serum albumin as standard, with two dilutions for each sample (l/10c and l/20e) and two determinations for each dilution. The mcan of these four concentrations was determined, and extracts were used only if the standard dcviation (SD) was less than 10%.
Distinct oligonucleotides containing complemcntary se-quences at their 3' termini and binding motifs for NF-kB were annealed and the protruding single-stranded regions filled in with (a-32P) dCTP (Amersham Pharmacia Biotech, Orsay, France) using Klenow fragment of DNA polymerase I (Promega, Charbonnieres, France). The sequences of the oligonucleotides used were as follows: 5'-AGTTGAGGCL GAC3TT-3'; 3'-CęCTGAAAGGGTCCG-5/.
The binding reaction mixture (10 pL) for gel retardation assays contained 1 ng of DNA probc, 2 pg poly'dI-dC, and 5 pg of the different nuclear protein extracts. The mixturc was ineubated for 30 min at 4°C. The DNA protein com-plexes were resoWed in native 5% polyacrylamide gels in 0.5 X Tris-borate ethylcnediaminetetraacetic acid (EDTA) at 100 V for 45 min. The dried gels were exposcd to Trimax
Wyszukiwarka
Podobne podstrony:
NF-kB and radiosensilivity • C. Didelot et al. 135712 3 412
img086 Chengjang Shimeid and Holland 2000Myllokunmingia fengjiaoa Shu et al., 1999 - &nb
14 H. Pridalova et al.INTRODUCTION Goats are bred worldwide and therefore there is a great variance
18 H. Pridalova et al. Table 3. Average values for goat cheeses - physical and Chemical properties T
29 F.SIMARD ET AL Exlruction and isolation. The air-dried (.1 wcek) pow-dered wood froni P. resinosa
© 302 PARKZ<FRA6E and Afadin in breast cancer A Letessier et al n
304 PARKZ>FRA6E and Afadin in breast cancer A Letessia et al Fijiurc 5 Afadin knockdown in MDCKIl
305 PARK2/FRA6E and Afadin in breast cancer A Letessier et al FRA6EJPARK2. The multivariate analysis
299 PARK2/FRA6E and Afadin in breast cancer A letessier et al lines can be suppressed by microcell-m
300 PARK2<FRA6E and Afadin in breast cancer A Letessief et al A 6q26-<*27 B
F00574 001 f003 Normal rangę © Elsevier. Boon et al.: Davidson s Principles and Practice of Medici
F00574 001 f006 The gap/blockj—ii—O Priority-setting © Elsevier. Boon et al.: Davidson s Principles
F00574 003 f003 Cłuiescent incomplete DNA repair © Elsevier. Boon et al.: Davidson
F00574 016 f007 tKł intake © Elsevier. Boon et al.: Davidson s Principles and Practice of Medicine 2
F00574 017 f035a [a] Normal tubular histołogy © Elsevier. Boon et al.: Davidson s Principles and Pra
F00574 017 f035c [Ć] Acute pyelonephritis © Elsevier. Boon et al.: Davidson s Principles and Practic
F00574 018 f032 Send or go for help as soon as possible © Elsevier. Boon et al.: Davidson s Principl
więcej podobnych podstron