Jacek Kluska
Department of Electrical and Computer Engineering
Rzeszow University of Technology, Poland
Application of data classifiers to
breast cancer diagnosis

Contents
2
Breast cancer input data
Overview of data classifiers
Exponential Radial Basis Function SVM (ERBFSVM)
Radial Basis Function Neural Network (RBFNN)
Learning Vector Quantization Neural Network (LVQNN)
Two Layer Perceptron (2LP)
Back Propagation Network (BPNN)
Decision supporting parameters
Cross Validation error
Diagnostic accuracy measures
Comparative analysis
Conclusions

Introduction
3
The goal:
Comparative analysis of ANNs and SVM in the
classification of medical data: breast cancer.
The input space is represented by the population of
683 women who suffered from breast cancer with 10
attributes associated with each patient.
Two data set classification problem, since each
instance has one of two possible cases: benign or
malignant.
In order to find which of the models predicts unknown
data more accurately, a 10-fold Cross Validation (CV)
algorithm is used.
Additionally, the diagnostic accuracy of the classifiers
is measured.

Introduction – cont.
4
Breast cancer input data (Wolberg, Mangasarian):
www.ics.uci.edu/~mlearn/databases/breast-cancer-
wisconsin/
Cardinality of learning data set
Total number of attributes
:
1.
clump thickness (1,2,…,10)
2.
uniformity of cell size (1,2,…,10)
3.
uniformity of cell shape (1,2,…,10)
4.
marginal adhesion (1,2,…,10)
5.
single epithelial cell size (1,2,…,10)
6.
bare nuclei (1,2,…,10)
7.
bland chromatin (1,2,…,10)
8.
normal nucleoli (1,2,…,10)
9.
mitoses (1,2,…,10)
9
n
683
l

ERBFSVM
5
ERBF kernel
Solution: optimal classifier
____________________________________
where
*
sign
,
i i
i
i SV
y
y K
b
x x
α
α
*
1
,
,
,
2
,
:
1,
1,
|
0
i i
i
r
i
s
i SV
r
s
i
b
y K
K
r s
SV y
y
SV
i
x x
x x
2
,
exp
2
i
i
K
x
x
x x
LVQNN
6
The output (1 neuron):
The outputs of the competitive hidden layer neurons
compet returns 1 only for the neuron whose weight vector
forms the closest match with the input x.
(2)
(1)
(1)
(1)
(1)
1
,
,
, ,
k
y
y
y
w
y
y
(1)
(1)
(1)
(1)
(1)
,
,1
compet
,
, ,
j
j
j
j n
j
y
w
w
x
w
w
RBF network
7
The output (1 neuron):
The outputs of hidden layer neurons
- spread constant
(2)
(1)
(1)
(1)
(1)
1
,
,
, ,
k
y
y
y
w
y
y
(1)
(1)
2
(1)
(1)
(1)
,
,1
exp
,
2( )
, ,
j
j
j
j n
j
y
sc
w
w
x
w
w
sc
2LP
8
The output (1 neuron):
where
The outputs of hidden layer neurons:
(2)
(1)
(2)
(1)
(1)
(1)
1
sign
,
, ,
k
y
b
y
y
w
y
y
(1)
(1)
(1)
sign
,
j
j
y
b
w
x
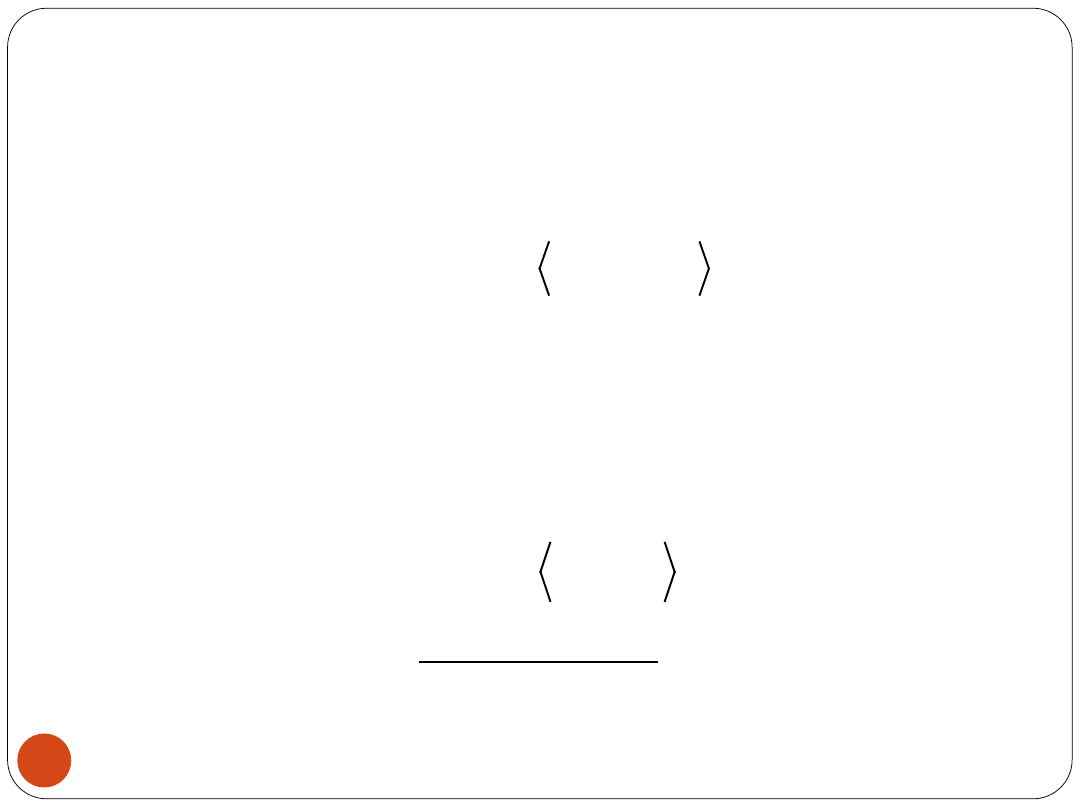
BPN
9
The output (1 neuron):
where
The outputs of hidden layer neurons:
(2)
(1)
(2)
(1)
(1)
(1)
1
logsig
,
, ,
k
y
b
y
y
w
y
y
(1)
(1)
(1)
logsig
,
1
logsig( )
,
0
1
exp
j
j
y
b
x
x
w
x
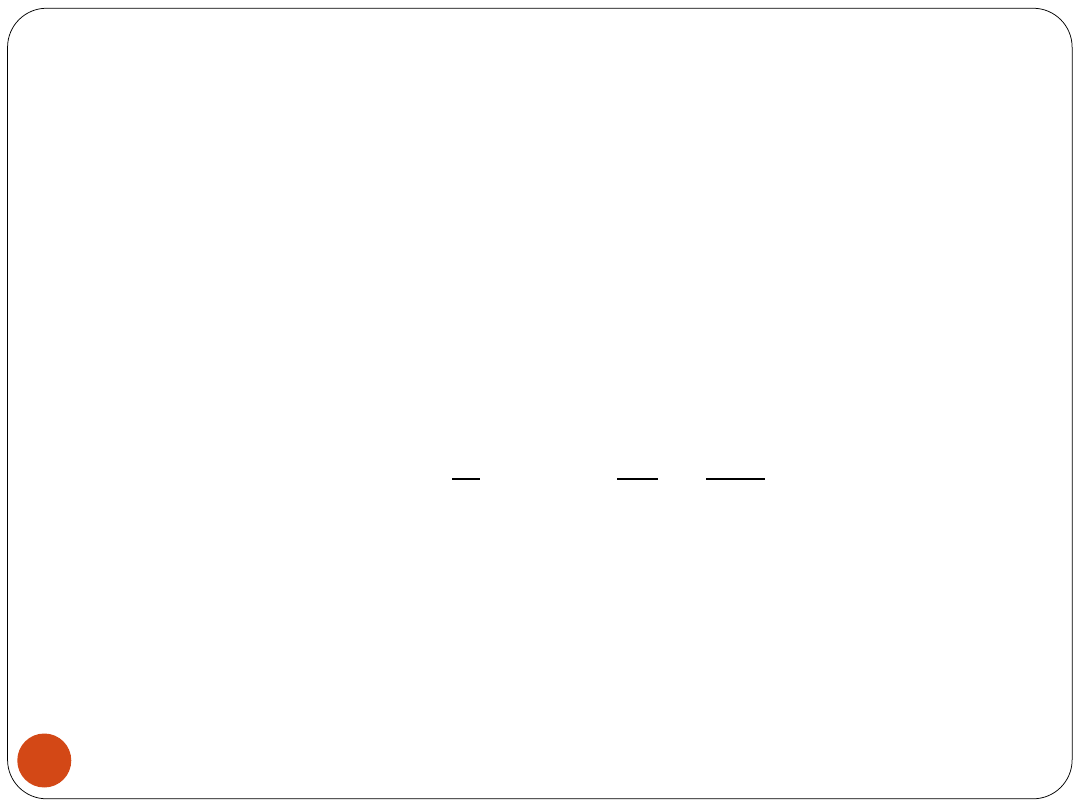
Cross Validation procedure
10
1.
Division of all input patterns (e.g. l = 199) into K
subsets, (e.g. K = 10).
2.
Model training on all subsets except from one.
3.
Model testing on the subset left out.
Cross Validation error (should be as low as
possible):
- the number of misclassified examples
within a single m
th
separation.
L - the number of validating data.
1
100
K
CV
i
i
E
L
L
K
m
i
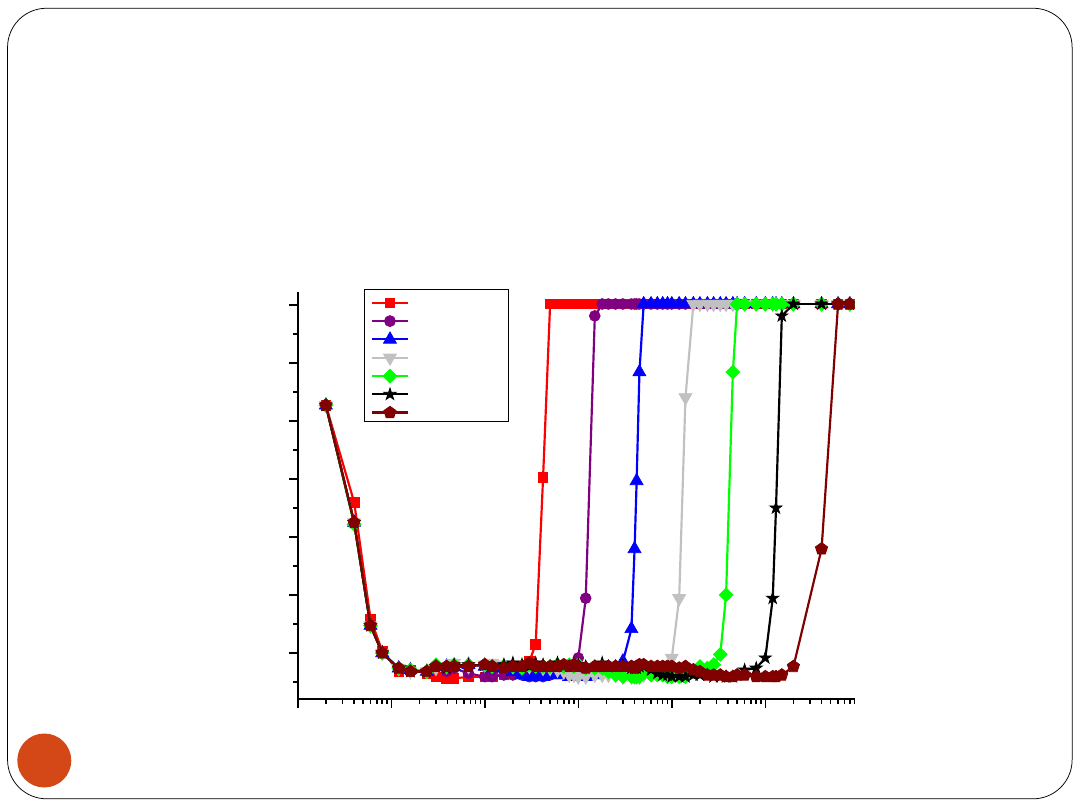
ERBFSVM results
11
The lowest percentage of misclassified examples:
E
CV
= 2.78 [%] by C = 1 and
σ ∈ {3.9,4.7}
0.1
1
10
100
1000
10000
5
10
15
20
25
30
35
E
CV
[%]
σ
C = 1
C = 10
C = 100
C = 1 000
C = 10 000
C = 100 000
C = 1 000 000
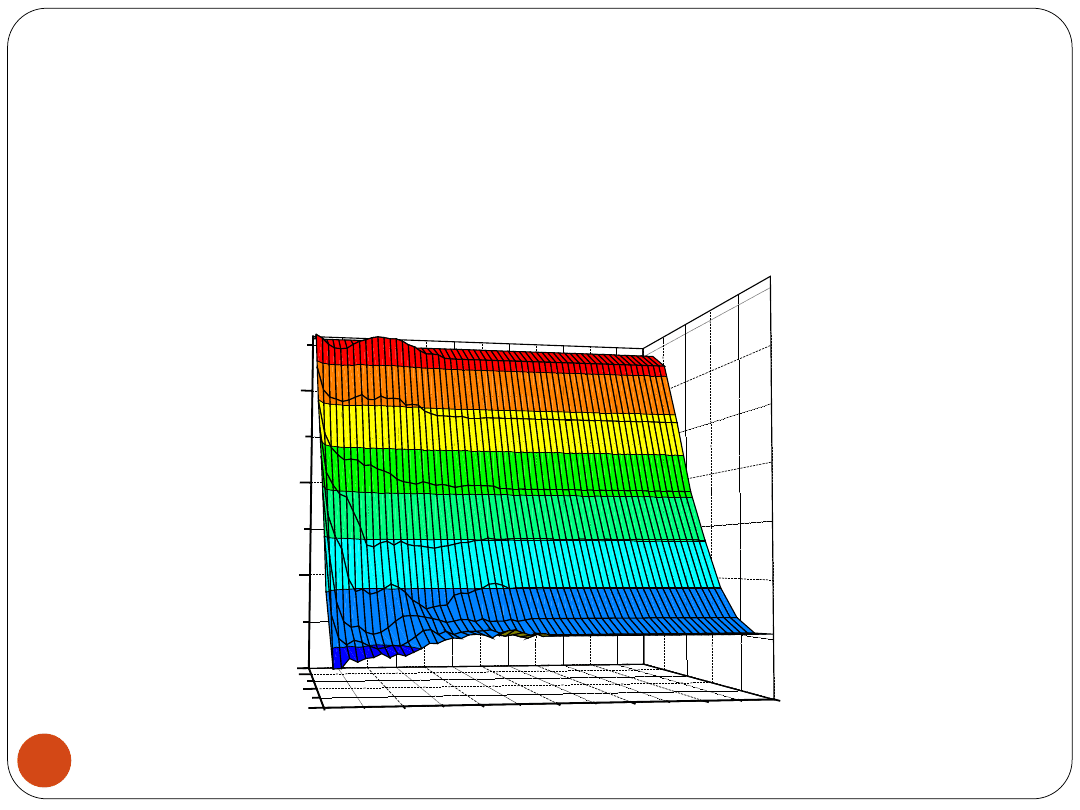
RBFNN results
12
The lowest percentage of misclassified examples:
E
CV
= 2.78 [%] by S1
∈ {9,11,13, ..., 29}
2
4
6
0
10
20
30
0
100
200
300
400
500
600
E
CV
[%]
S1
sc
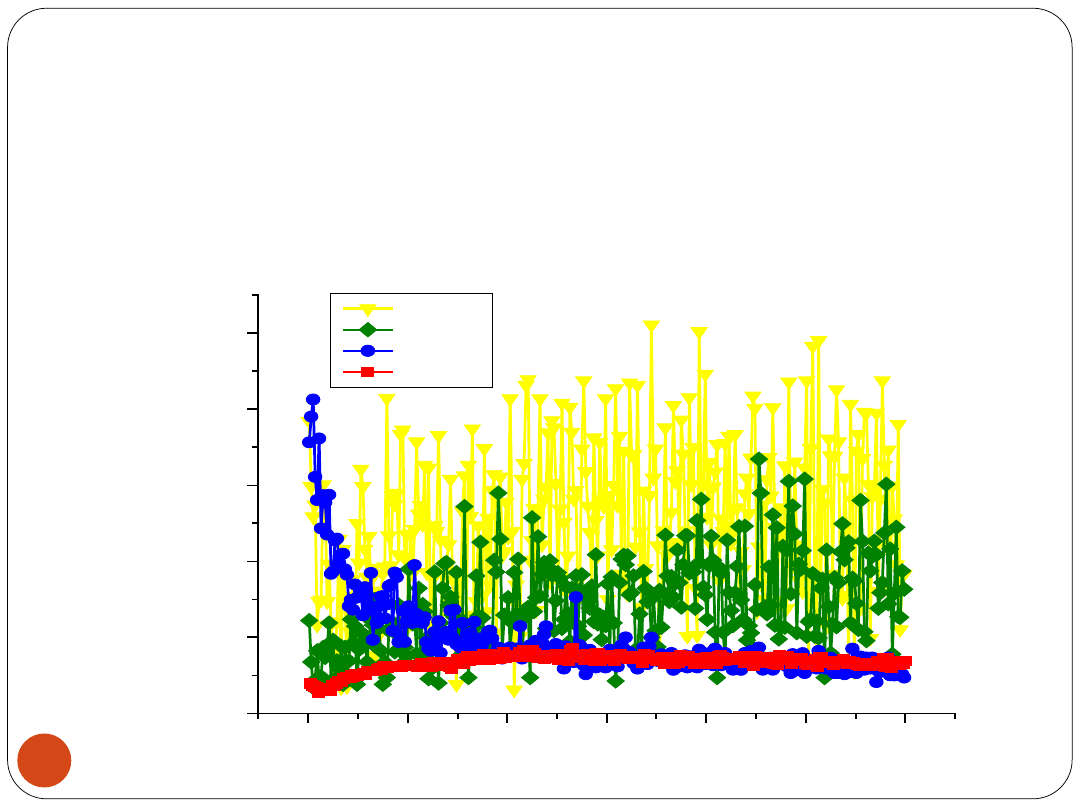
LVQNN vs. remaining networks
13
LVQNN accomplishes identical lowest prediction error:
ERBFSVM and RBFNN, E
CV
= 2.78 [%] by S1 = 10
0
100
200
300
400
500
600
0
10
20
30
40
50
E
CV
[%]
2TBPNN
2LBPNN
2LP
LVQNN
σ
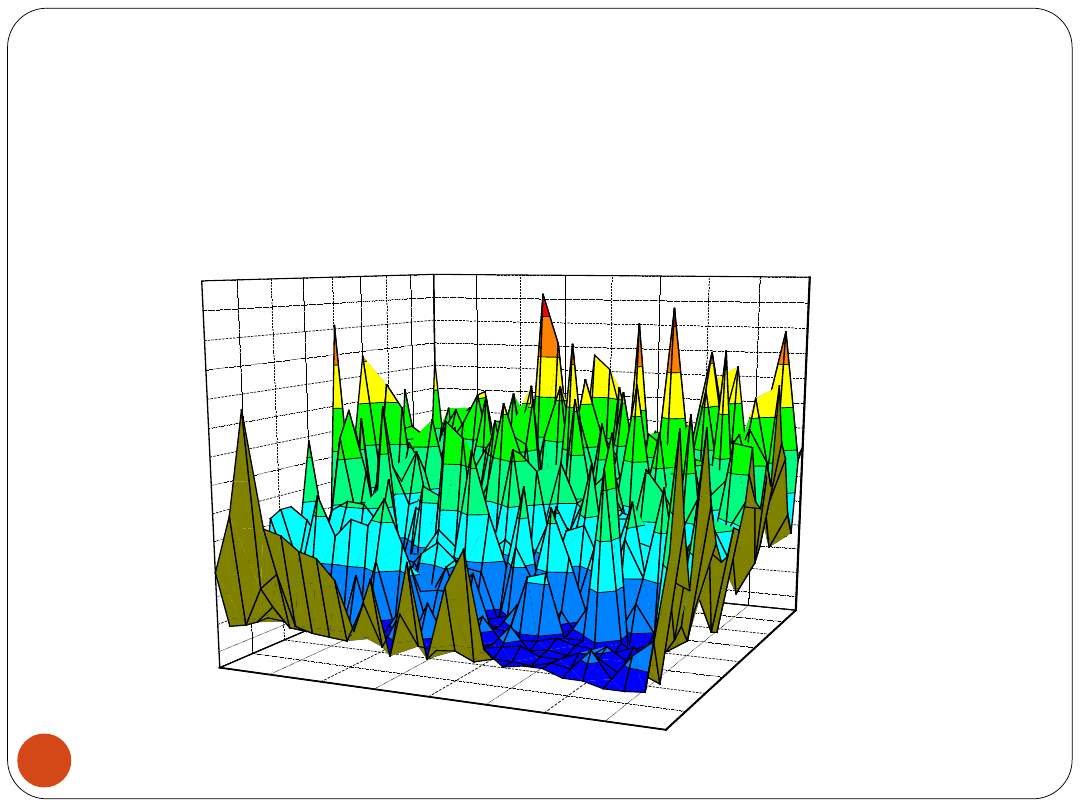
3LBPNN
14
3 layer BPNN is completely unpredictable as a
classifier ...
50
100
150
200
50
100
150
200
5
10
15
20
25
30
35
E
CV
[%]
S2
S1

Diagnostic accuracy
15
TruePos
sensitivity
TruePos
FalseNeg
TrueNeg
specificity
TrueNeg
FalsePos
16
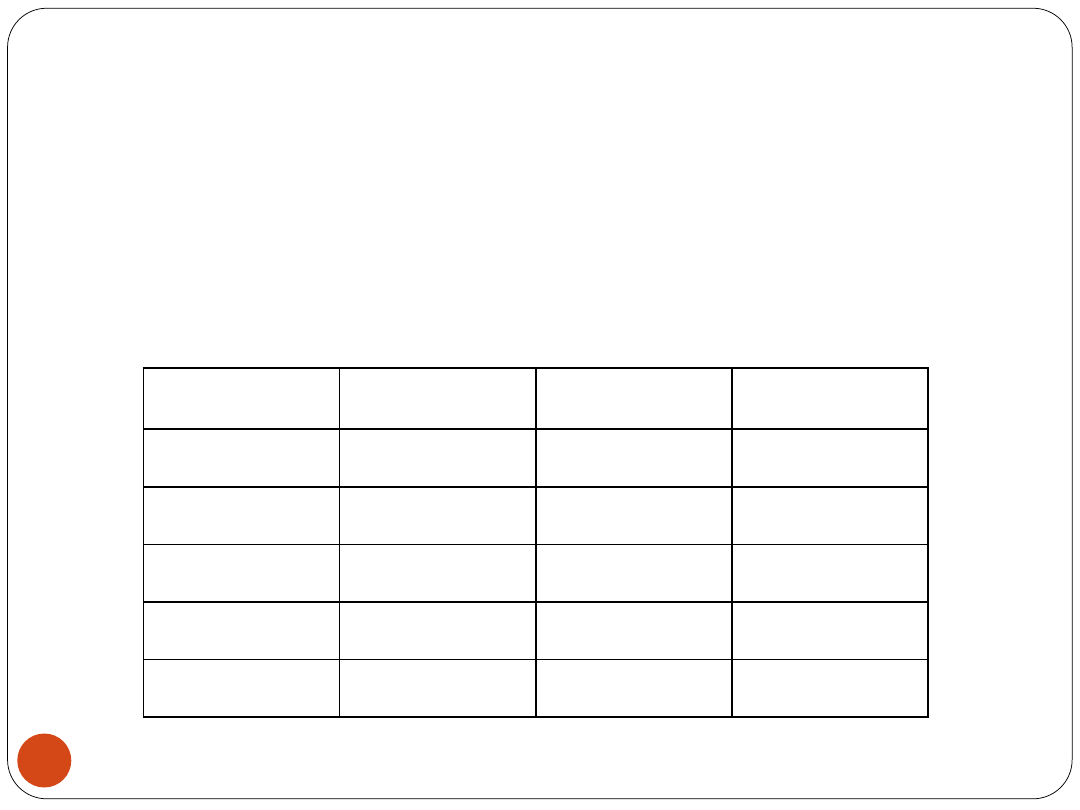
Generalization results –
summary
Classifier
E
CV
[%]
sensitivity
specificity
ERBSFM
2.78
0.95
0.98
RBFNN
2.78
0.95
0.98
LVQNN
2.78
0.95
0.98
2LP
4.11
0.93
0.94
3BPNN
3.53
0.93
0.95
Diagnostic accuracy and generalization
ability of breast cancer data classifiers

Conclusions
17
The sensitivity 0.95 for ERBFSVM, LVQNN and
RBFNN means that 95% of sick patients is identified
as sick.
Specificity 0.98 gives 98% of certainty that healthy
patients are diagnosed as such.
High diagnostic accuracy is justified by a very good
generalization ability of the models; E
CV
= 2.78 [%].
This suggests that these classifiers are reliable and
precise in medical diagnosis on new breast cancer
cases.
The above models can serve as a feedback for
physicians during the process of treatment.
Conclusions – cont.
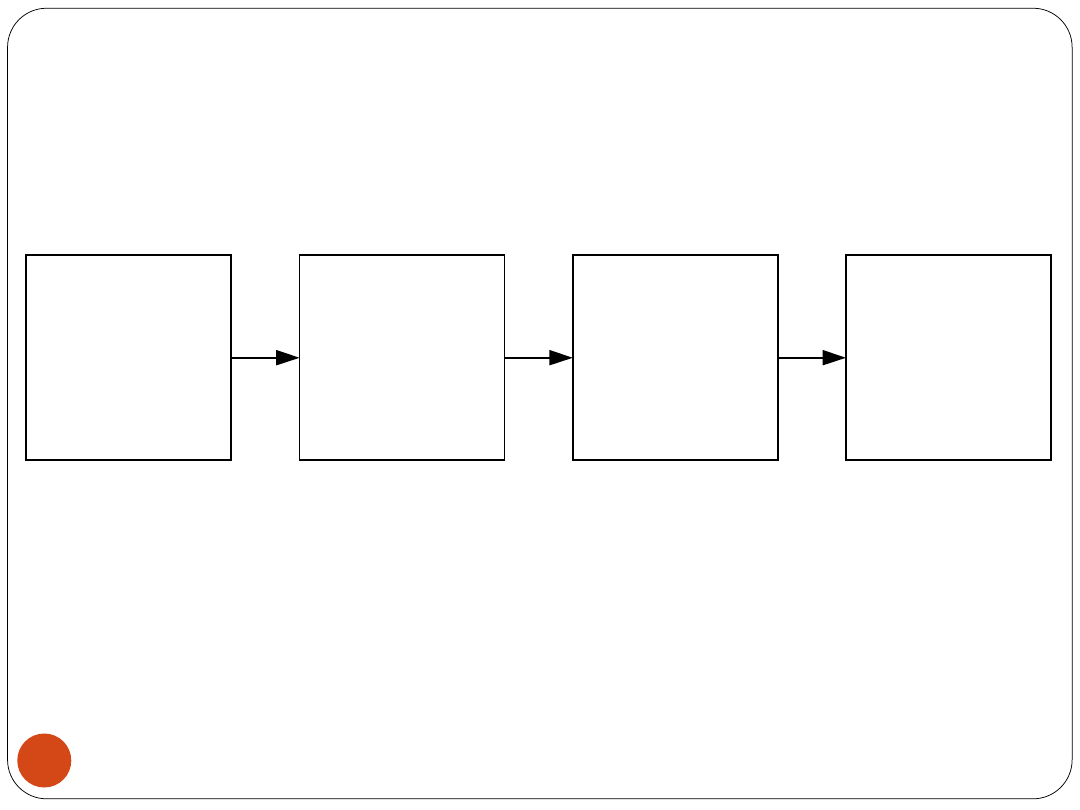
18
Medical Diagnostic System
Acknowledgements
The author is grateful to PhD student Maciej Kusy for his valuable help with data
preparation and calculations.
Module I
Data Preparation
Module III
Main Routine -
Training and
Testing of Models
Module II
Specification and
Initialization of
Models
Module IV
Selection of Most
Optimal Model.
Rule Generation
Document Outline
- Application of data classifiers to breast cancer diagnosis
- Contents
- Introduction
- Introduction – cont.
- ERBFSVM
- LVQNN
- RBF network
- 2LP
- BPN
- Cross Validation procedure
- ERBFSVM results
- RBFNN results
- LVQNN vs. remaining networks
- 3LBPNN
- Diagnostic accuracy
- Slide Number 16
- Conclusions
- Conclusions – cont.
Wyszukiwarka
Podobne podstrony:
Perceived risk and adherence to breast cancer screening guidelines
11 a Ovarian Cancer
2008 Coping With Breast Cancer Workbook for couples
The Relationship between Twenty Missense ATM Variants and Breast Cancer Risk The Multiethnic Cohort
A nonsense mutation (E1978X) in the ATM gene is associated with breast cancer
Missense Variants in ATM in 26,101 Breast Cancer Cases an 29,842 Controls
INTERNET USE AND SOCIAL SUPPORT IN WOMEN WITH BREAST CANCER
Evaluation of the role of Finnish ataxia telangiectasia mutations in hereditary predisposition to br
Population Based Estimates of Breast Cancer Risks Associated With ATM Gene Variants c 7271T4G and c
Quality of life of 5–10 year breast cancer survivors diagnosed between age 40 and 49
Established breast cancer risk factors by clinically important
Rare, Evolutionarily Unlikely Missense Substitutions in ATM Confer Increased Risk of Breast Cancer
03 Antibody conjugated magnetic PLGA nanoparticles for diagnosis and treatment of breast cancer
Single nucleotide polymorphism D1853N of the ATM gene may alter the risk for breast cancer
Variants in the ATM gene associated with a reduced risk of contralateral breast cancer
Breast Cancer Early Detection ACS
Spectrum of ATM Gene Mutations in a Hospital based Series of Unselected Breast Cancer Patients
Variants in the ATM gene and breast cancer susceptibility
Predictors of perceived breast cancer risk and the relation between preceived risk and breast cancer
więcej podobnych podstron