
ICIP-2002 Paper Proposal:
CONTENT-ADAPTIVE MESH MODELING FOR FULLY-3D
TOMOGRAPHIC IMAGE RECONSTRUCTION
1
Yongyi Yang, Jovan G. Brankov, and Miles N. Wernick
Dept. of Electrical and Computer Engineering
Illinois Institute of Technology
3301 S. Dearborn St., Chicago, IL 60616, USA
1
This research was supported by the Whitaker Foundation and by NIH/NHLBI grant HL65425.
ABSTRACT
In this paper we propose the use of a content-adaptive
volumetric mesh model for fully three-dimensional (3D)
tomographic image reconstruction. In the proposed
framework, the image to be reconstructed is first modeled
by an efficient mesh representation. The image is then
obtained through estimation of the nodal values from the
measured data. The use of a mesh representation can
alleviate the ill-posed nature of the reconstruction
problem, thereby leading to improved quality in the
reconstructed images. In addition, it reduces the data
storage requirement, resulting in efficient algorithms. The
proposed methods are tested using gated cardiac-
perfusion images. Initial results demonstrate that the
proposed approach achieves superior performance when
compared to several commonly used methods for image
reconstruction, and produces results very rapidly.
1. INTRODUCTION
In recent years there has been growing interest in fully-3D
tomographic image reconstruction. A major challenge in
fully-3D reconstruction lies in its memory requirement
and demanding computation time. Like their 2D
counterpart, most 3D reconstruction methods have
traditionally been developed based on voxel image
representations [1]. Bayesian priors (e.g., [2]) or
regularization terms (e.g., [3]) are typically used to
combat the effect of noise.
Alternative model-based reconstruction approaches
have also been proposed. For example, cylindrical models
were proposed in [4] and surface models were used in
[5,6].
In our previous work in [7], a content-adaptive mesh
modeling approach was proposed for 2D image
reconstruction. It was demonstrated that such an approach
can outperform several well-known reconstruction
algorithms in terms of both reconstructed image quality
and computation time. In this study, we extend this
approach to fully-3D image reconstruction. In this new
approach, the image is first modeled by a volumetric mesh
model, on the basis of which a customized basis
representation is obtained for the image. The parameters
of this representation are then estimated from the data.
In a volumetric mesh model, the 3D image domain is
subdivided into a collection of mesh elements, the vertices
of which are called nodes. The image function is then
obtained over each element by interpolation from the
values of these nodes [8]. In a content-adaptive mesh
model (CAMM), the mesh elements are placed in a
fashion that is adapted to the local content of the image. A
CAMM provides an efficient representation of the image
in that the number of parameters (i.e., mesh nodes) is
typically much less than the number of required voxels in
a voxel image representation. In addition, a mesh model
can also be used for motion tracking in an image
sequence, by allowing the mesh to deform over time [9].
The potential benefits of using a CAMM for image
reconstruction are: 1) a CAMM reduces the number of
unknowns, thus alleviating both the underdetermined
nature of the reconstruction problem and the data storage
requirement, particularly for the case of 3D
reconstruction; 2) this reduction in the number of
unknowns can also lead to a fast computation; 3) a
CAMM provides a natural spatially-adaptive smoothness
mechanism; and 4) the CAMM provides a natural
framework for reconstruction of moving image sequences.
2. METHODS
2.1 Mesh Tomography Model
Let f x
a f
denote the image function defined over a domain
D
, which is 3D in this study. In a mesh model, the domain
D
is partitioned into
M
non-overlapping mesh elements,
denoted by
,
1, 2, ,
m
D
m
M
=
"
. The image function is
represented as
( )
( ) ( )
1
( )
N
n
n
n
f
f
e
ϕ
=
=
+
∑
x
x
x
x
,
(1)
where
x
n
is the nth mesh node,
ϕ
n
x
a f
is the interpolation
basis function associated with
x
n
, N is the total number
of mesh nodes used, and
( )
e
x
is the modeling error. Note
that the support of each basis function
ϕ
n
x
a f
is limited to
those elements
D
m
attached to the node
n
. In this study,
tetrahedrons are used for
D
m
, and linear interpolation
functions are used for
( )
n
ϕ
x
.
Now let
n denote a vector formed by the nodal values
of the mesh model, i.e.,
( ) ( )
( )
1
2
,
,
T
n
f
f
f
≡
n
x
x
x
"
. (2)
If
f
denotes the voxel representation of the image function
f
x
a f
over
D
, then from (1) and (2) one can obtain
= Φ +
f
n e
, (3)
where
Φ
is a matrix, composed from the interpolation
functions
ϕ
n
x
a f
in (1), that forms the interpolation
operator from a mesh representation to the pixel
representation, and e is a vector denoting the error
( )
e
x
.
For tomographic image reconstruction, the imaging
equation is typically written in terms of the voxel
representation
f
as
E[ ]
g
Hf
=
, (4)
where
g contains the measured data,
E[ ]
⋅
is the
expectation operator, and
H
is a matrix describing the
imaging system.
Substituting (3) into (4), we obtain the mesh-domain
imaging equation:
[
]
ˆ
[ ]
E
=
Φ + ≡
+
g
H
n e
An e
, (5)
where
ˆ
, and .
= Φ
=
A H
e He
As demonstrated later, a CAMM can provide a very
accurate representation of the original image. As a result,
the modeling error ˆ
e
in (5) can be ignored when
compared to the noise level in the imaging data. Thus, we
have
[ ]
E
≈
g
An
. (6)
The reconstruction problem becomes that of estimating
n
from the observed data
g . The image
f
can then be
obtained from (3) (with
e ignored).
2.2 Reconstruction Algorithms
In this paper we investigate maximum-likelihood and
least-squares estimates of the nodal values in
n
.
A. Maximum-Likelihood Estimate
The maximum-likelihood (ML) estimate is obtained as
( )
{
}
ˆ
arg max log
;
ML
p
=
n
n
g n
, (7)
where
( )
;
p g n
is the likelihood function of
g
parameterized by
n
. In this paper, we assume a Poisson
likelihood, which characterizes emission tomography
The ML estimate can be computed by using the
following expectation-maximization (EM) algorithm [10]:
( )
(
1)
( )
j
j
s
t
s
ts
j
t
ts
tk
k
t
k
+
=
∑
∑
∑
n
g
n
A
A
A n
, (8)
where
( )
k
s
n
is the value of node
s in iteration j, g
t
is the
recorded count for observation
t
, and A
ts
is the
ts entry of
matrix A.
B. Least-Squares Estimate
The least-squares estimate is obtained as the solution of
the following optimization problem:
2
ˆ
arg min
LS
=
−
n
n
g An
, (9)
where
⋅ is the Euclidean norm. This quadratic objective
function has a unique solution, provided that A is of full
rank. In this study, we used the conjugate gradient
algorithm [11] to perform the optimization.
3. PRELIMINARY RESULTS
3.1 Evaluation Image Data
To demonstrate the proposed CAMM-based
reconstruction approach, we used the 4D gated
mathematical cardiac-torso (gMCAT) D1.01 phantom
[12], which is a time sequence of 16 3D images. The field
of view was 36 cm; the pixel size was 5.625mm. Poisson
noise, at a level of 4 million total counts per 3D time-
frame image, was introduced into the projections to
simulate a clinical
Tc
m
99
study. No attenuation correction
was used.
3.2 Volumetric Mesh Generation
The key to the proposed approach lies in how to
construct a CAMM that is compact and accurate for
representing the volumetric image to be reconstructed. For
this purpose we extended our method in [13] to the 3D
case. This method consists of the following three steps: 1)
extract a feature map
( )
σ x from the image
( )
f
x
based
on the largest magnitude of its second directional
directives; 2) apply the well-known Floyd-Steinberg error-
diffusion algorithm, a method originally designed for
digital halftoning [14], to distribute mesh nodes non-
uniformly in the 3D image domain, with density
proportional to the feature map
σ p
b g
; and 3) use a 3D
Delaunay triangulation algorithm [15] to connect the mesh
nodes. The resulting mesh consists of tetrahedral elements
that are automatically adapted to the content of the image.
To demonstrate the accuracy of the CAMM produced
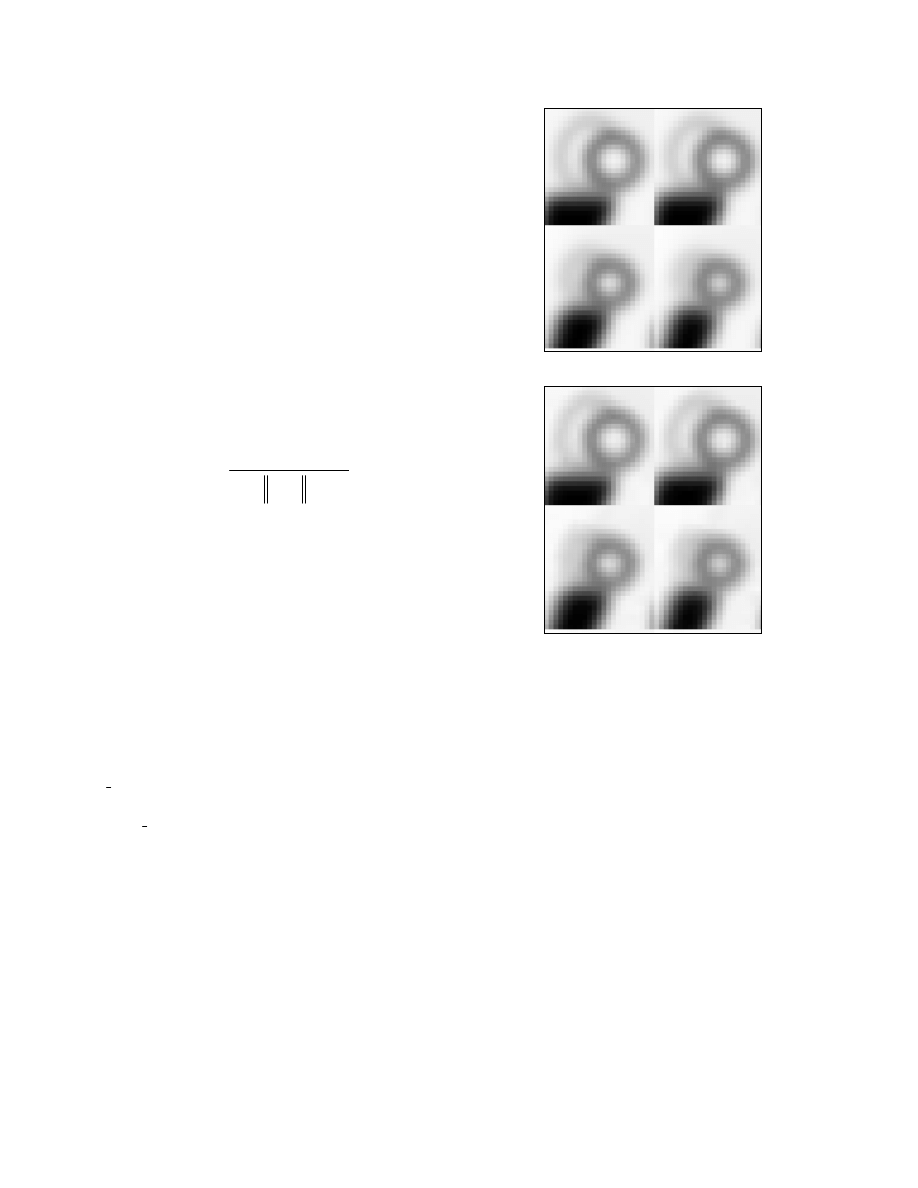
by this algorithm, we show in Fig. 1 some results obtained
for a 3D frame from the phantom. Shown in Fig. 1(a) is a
short-axis view of four slices selected in the vicinity of the
heart from this original volumetric frame. Shown in
Fig.1(b) are these same four slices from a mesh
representation of this volume obtained from our
algorithm, in which 10,688 mesh nodes were used (only
about 4% of the total number of voxels used in (a)). To
quantify the accuracy of this mesh representation, we
computed its peak-signal-to-noise ratio (PSNR) to be 42.8
dB. The PSNR is defined as
PSNR
M N L f
=
× × ⋅
−
F
H
GG
I
K
JJ
10
2
2
log
max
f f
dB
, (6)
where
f
and
f
denote the original image and its mesh
representation, respectively,
f
max
is the image peak value,
and
M N L
× ×
is the image dimension.
These results demonstrate that the proposed mesh
model can indeed provide an accurate representation of
the volumetric image with a very small number of mesh
nodes.
Of course, for tomographic image reconstruction the
mesh structure has to be estimated from the observed data.
The following procedure was demonstrated to work well
in our previous studies [7]. First, the projection data are
summed over the 16 gated frames. From these summed
projections an image is reconstructed using the filtered
back projection algorithm. The resulting image, denoted
by
f
x
a f
, provides a rough estimate of the heart summed
over all 16 frames. The mesh structure is then created
based on
f
x
a f
using the steps described above.
In Fig. 2 we show the mesh structure for a 2D slice
obtained from the projection data. The mesh nodes are
highlighted in Fig. 2 using bright dots. It is evident that
the obtained mesh structure is well adapted to the content
of the image, where mesh nodes have been automatically
placed densely in the important heart regions, and sparsely
in the background.
For better visualization purposes, additional results and
animations are provided for the obtained 3D mesh
structures at the following web site:
http://www.ipl.iit.edu/brankov/Rotate.htm. These results
(a) Original (262,144 voxels)
(b) CAMM (10,688 nodes)
Figure 1. (a) Short axis view of four slices selected in the
vicinity of the heart from an original 3D time-frame
(262,144 voxels) of the gMCAT phantom; and (b) the
same four slices in a mesh representation obtained by our
algorithm, in which only 10,688 mesh nodes were used.
The mesh representation has PSNR=42.8 dB.
demonstrate that the mesh structure produced by the
proposed method is well adapted to the content of the 3D
volumetric images.
3.3 Fully 3D CAMM Reconstruction
For validating the concept of the proposed CAMM
based reconstruction methods, our initial work has been
focused on using 2D models. These preliminary results
indicate that the proposed method can outperform several
existing pixel-based methods. We are currently applying
the volumetric mesh model described above to fully 3D
reconstruction.
Given the success we had using the 2D model, we
expect that the use of a fully 3D CAMM could offer even

Figure 2. Content-adaptive mesh model of the torso,
including the heart.
greater advantage for image reconstruction. This is
because a 3D CAMM can further exploit the redundancy
among the different 2D slices in a volumetric frame,
offering a much more compact representation than in the
2D case. This point is clearly demonstrated by the results
in Fig. 1, where the number of mesh nodes used was only
4% of the number of voxels in the original volumetric
frame, yet the mesh representation achieves a mean-
square-error as low as
5
5.25 10
−
×
(assuming image
dynamic range between 0 and 1). It is expected that such a
great reduction in the number of unknowns by the mesh
model would eventually lead to a very efficient
reconstruction algorithm. We plan to furnish detailed,
complete results of this fully 3D reconstruction method,
along with comparisons to other methods [e.g., 16-18], by
the time of the conference.
5. REFERENCES
[1] B. Bendriem and D. W. Townsend,
The Theory and
Practice of 3D PET, Kluwer Academic Publishers,
1998.
[2] Geman and D. Geman, “Stohastic relaxation, Gibbs
distributions, and Bayesian restoration of images,”
IEEE Trans. Patt. Anal. Mach. Intell., vol. 6, pp. 721-
741, 1984.
[3]
J. Fessler, “Penalized weighted least-squares
reconstruction for positron emission tomography,”
IEEE Trans. Med. Imaging, vol. 13, pp. 290-300, 1994.
[4] Y. Bresler, J. A. Fessler, and A. Macovski, “A
Bayesian approach to reconstruction form incomplete
projections of a multiple object 3D domains,”
IEEE
Trans. Patt. Anal. Mach. Intell., vol. 11, pp. 840-858,
1989.
[5] G. S. Cunningham, K. M. Hanson, and X. L. Battle,
“Three dimensional reconstruction from low-count
SPECT data using deformable models,"
IEEE. Med.
Imaging Conf., 1997.
[6] G. R. Jennings and D. R. Wolf, “Tomographic
reconstruction based on flexible geometric models,”
IEEE Int. Conf. on Image Proc., 1994.
[7] J. G. Brankov, Y. Yang, and M. N. Wernick,
“Tomographic image reconstruction using content-
adaptive mesh modeling,”
IEEE Inter. Conf. Image
Proc., vol. 1, pp. 690-693, Thessaloniki, Greece, Oct.,
2001.
[8] Y. Wang and L. O., “Use of two-dimensional
deformable mesh structures for video coding--I: The
synthesis problem: mesh-based function approximation
and mapping,”
IEEE Trans. Circuits Syst. for Video
Tech., vol. 6, pp. 636 -646, 1996.
[9] Y. Altunbasak and A. M. Tekalp, “Occlusion-
adaptive, content-based mesh design and forward
tracking,”
IEEE Trans. Image Proc., vol. 6, pp. 1270-
1280, 1997.
[10] A. P. Dempster, N. M. Laird, and D. B. Rubin,
“Maximum likelihood from incomplete data via the EM
algorithm,”
J. Roy. Statist. Sect., vol. 39, pp. 1-38,
1977.
[11] E. K. P. Chong and S. H. Zak,
An Introduction to
Optimization. New York: John Wiley & Sons, Inc.,
1996.
[12] P. H. Pretorius, M. A. K. W. Xia, B. M. W. Tsui, T.
S. Pan, and B. J. Villegas, “Evaluation of right and left
ventricular volume and ejection fraction using a
mathematical cardiac torso phantom for gated blood
pool SPECT,”
J. of Nucl. Med, vol. 38, pp. 1528-1534,
1997.
[13] Y. Yang, J. Brankov, and M. N. Wernick, “A fast
algorithm for accurate content-adaptive mesh
generation,”
IEEE Inter. Conf. Image Proc., vol. 1, pp.
868-871, Thessaloniki, Greece, Oct., 2001.
[14] R. Floyd and L. Steinberg, “An adaptive algorithm
for spatial gray scale,” SID Int. Sym. Digest of Tech.
Papers, 1975.
[15] F. Preparata and M. Shamos,
Computational
Geometry--An Introduction, Springer-Verlag, New
York, 1985.
[16] D. S. Lalush and B. M. W. Tsui, “Space-Time Gibbs
Priors applied to Gated SPECT Myocardial Perfusion
Studies,” presented at
3D Image Rec. in Radiology,
Dordrecht, 1996.
[17] H. Hudson and R. Larkin, “Accelerated image
reconstruction using ordered subsets of projection
data,”
IEEE Trans Med Imag, Vol.13, pp.601-609,
1994.
[18] H. Erdogan and J. A. Fessler, “Ordered subsets
algorithms for transmission tomography,”
Phys. Med.
Biol, Vol.44, 1999.
Wyszukiwarka
Podobne podstrony:
Adaptive fuzzy control for uninterruptible power supply with three phase PWM inverter
Adaptive fuzzy control for uninterruptible power supply with three phase PWM inverter
Going 3D Survival Guide for 2D CAD Users
Modeling Of The Wind Turbine With A Doubly Fed Induction Generator For Grid Integration Studies
Adapting Quick?itor for COM
Rational UML Profile for business modeling IBM
Bangia, Diebold, Schuermann And Stroughair Modeling Liquidity Risk, With Implications For Traditiona
MODELING OF THE ACOUSTO ELECTROMAGNETIC METHOD FOR IONOSPHERE MONITORING EP 32(0275)
A MODAL ARGUMENT FOR NARROW CONTENT
On the Infeasibility of Modeling Polymorphic Shellcode for Signature Detection
BSF content for test one
using uml for modeling a distributed java application 1997
AES Information Document For Room Acoustics And Sound Reinforcement Systems Loudspeaker Modeling An
guidelines for the content of rig move procedures sept 2008
Process Modeling, Simulation, and Control for Chemical Engineers 2E
więcej podobnych podstron