
On the frequency of protein glycosylation, as deduced from analysis of
the SWISS-PROT database
1
Rolf Apweiler
a;
*, Henning Hermjakob
a
, Nathan Sharon
b
a
EMBL Outstation Hinxton, European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, UK
b
Department of Membrane Research and Biophysics, The Weizmann Institute of Science, Rehovot 76100, Israel
Received 15 February 1999; accepted 19 May 1999
Abstract
The SWISS-PROT protein sequence data bank contains at present nearly 75 000 entries, almost two thirds of which
include the potential N-glycosylation consensus sequence, or sequon, NXS/T (where X can be any amino acid but proline)
and thus may be glycoproteins. The number of proteins filed as glycoproteins is however considerably smaller, 7942, of which
749 have been characterized with respect to the total number of their carbohydrate units and sites of attachment of the latter
to the protein, as well as the nature of the carbohydrate-peptide linking group. Of these well characterized glycoproteins,
about 90% carry either N-linked carbohydrate units alone or both N- and O-linked ones, attached at 1297 N-glycosylation
sites (1.9 per glycoprotein molecule) and the rest are O-glycosylated only. Since the total number of sequons in the well
characterized glycoproteins is 1968, their rate of occupancy is 2/3. Assuming that the same number of N-linked units and rate
of sequon occupancy occur in all sequon containing proteins and that the proportion of solely O-glycosylated proteins (ca.
10%) will also be the same as among the well characterized ones, we conclude that the majority of sequon containing proteins
will be found to be glycosylated and that more than half of all proteins are glycoproteins. ß 1999 Elsevier Science B.V. All
rights reserved.
Keywords: Glycosylation; Glycoprotein; Database
Glycosylation is a common and highly diverse co-
and post-translational protein modi¢cation reaction.
Perhaps because almost all proteins of human serum
and of hen egg-white are glycosylated [1], as are
those of animal cell membranes [2], the sweeping
statement has been made that `most proteins are gly-
coproteins' [3,4]. The recent development of compu-
terized protein sequence data banks allows us to put
this statement to a quantitative test. Here, we present
the results of such an attempt, based on an analysis
of the SWISS-PROT database [5]. This data bank is
manually curated and strives to provide a high qual-
ity of annotation, with a minimal level of redun-
dancy and a high level of integration with other bio-
molecular databases. It is thus more reliable than the
supplementary computer-annotated TrEMBL data
bank that contains translations of all protein coding
sequences in the EMBL nucleotide sequence data-
base which are not yet in the SWISS-PROT and is
therefore much larger than the latter.
In almost all glycoproteins, the carbohydrate units
are attached to the protein backbone either by N- or
O-glycosidic bonds or by both types of linkage. The
0304-4165 / 99 / $ ^ see front matter ß 1999 Elsevier Science B.V. All rights reserved.
PII: S 0 3 0 4 - 4 1 6 5 ( 9 9 ) 0 0 1 6 5 - 8
* Corresponding author..
1
Dedicated to Prof. Akira Kobata and Prof. Harry Schachter
on the occasion of their 65th birthdays.
BBAGEN 24913 16-11-99
Biochimica et Biophysica Acta 1473 (1999) 4^8
www.elsevier.com/locate/bba
N-glycosidic bond is always to the amide of an as-
paragine that is part of the consensus sequence NXS/
T, or `sequon', where X can be any amino acid ex-
cept proline. The sequons are often referred to as
`potential glycosylation sites', since, for reasons that
are not understood, in not all of these, the aspara-
gine is glycosylated. No consensus sequences for
O-glycosylation seem to exist.
The SWISS-PROT database contained by the end
of 1998 (release 36, including updates to 01/11/98)
74 988 entries. Potential N-glycosylation sites were
identi¢ed 151 993 times in 48 636 sequences, an aver-
age of 3.1 per protein. In 26 352 protein sequences,
such sites are absent, showing that about one third of
the proteins cannot be N-glycosylated. Examination
of the TrEMBL entries leads to similar conclusions
(Table 1).
The number of proteins in SWISS-PROT that
have been ¢led as glycoproteins is relatively small,
namely 7942 (10.6% of the total). In this database,
a protein is labelled with the keyword `GLYCOPRO-
TEIN' only when it is beyond reasonable doubt
that it is really glycosylated, even if information on
the nature of its carbohydrate units and their linkage
to the protein is lacking, as is the case for most of
these glycoproteins. Detailed annotation on the gly-
cosylation of these substances is done by the follow-
ing format: `FT CARBOHYD 6 position s 6 po-
Table 1
Protein entries and sequons in SWISS-PROT and TrEMBL da-
tabases by the end of 1998
Database
SWISS-PROT TrEMBL
Number of entries
74 988
156 187
Entries containing NXS/T sequon
48 636
107 551
64.9%
68.9%
Number of sequons
151 933
394 483
Sequon/sequon containing entry
3.12
3.66
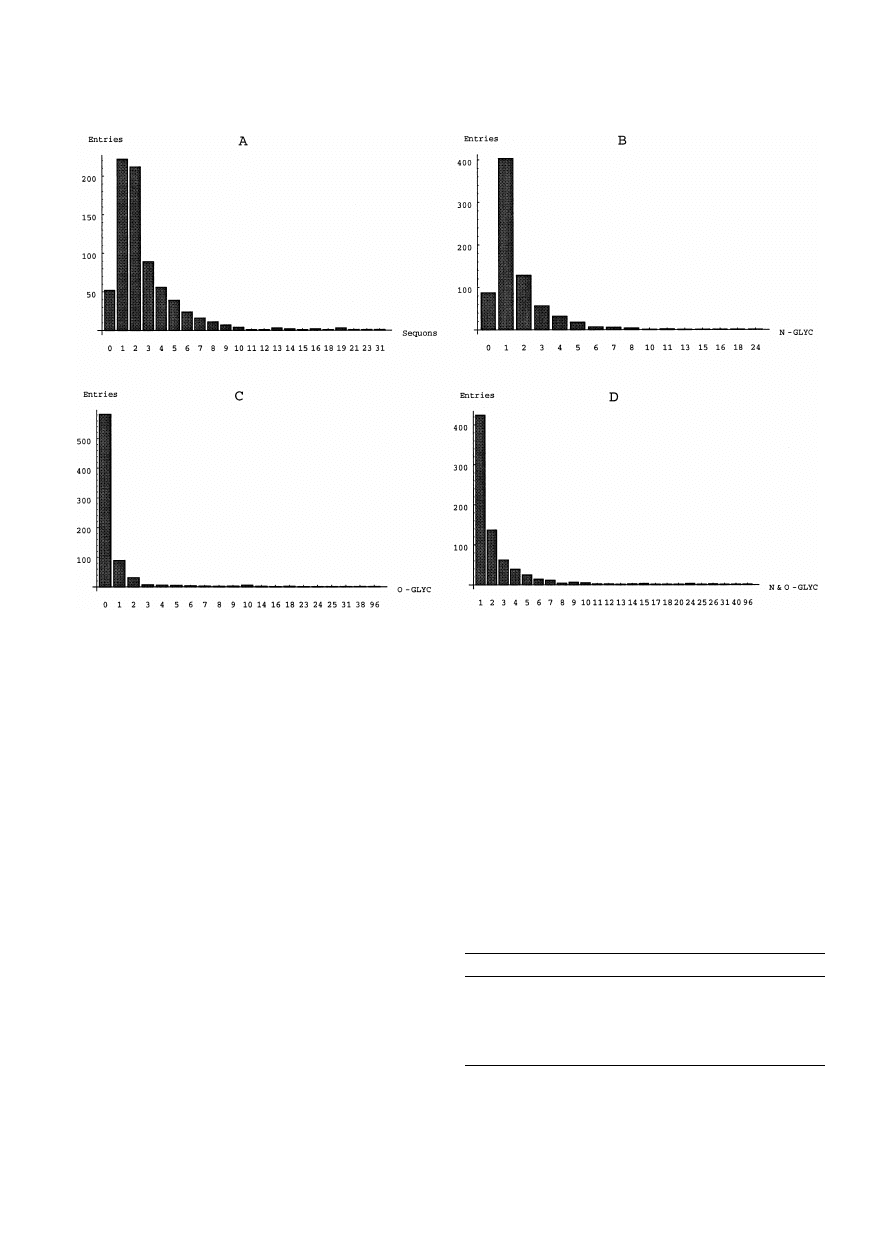
Fig. 1. Frequency of occurrence of sequons and carbohydrate units in the 749 well characterized glycoproteins listed in the SWISS-
PROT database by the end of 1998. Sequons per glycoprotein (A), and carbohydrate units in N-glycoproteins (B), O-glycoproteins
(C) and N-,O-glycoproteins (D).
BBAGEN 24913 16-11-99
R. Apweiler et al. / Biochimica et Biophysica Acta 1473 (1999) 4^8
5
sition s 6 description s '. For example, a glycopro-
tein with a N-acetylgalactosamine at position 34 will
be annotated as `FT CARBOHYD 34 34 N-ACE-
TYLGALACTOSAMINE'.
As a thorough biochemical characterization of
most of the glycoproteins is lacking, numerous 6 de-
scription s tags contain the strings `BY SIMILAR-
ITY', `PROBABLE' or `POTENTIAL'. These de-
note that no biochemical characterization of the
glycosylation site(s) is available. For the purpose of
Table 2
Potential and real glycosylation sites in the 749 well characterized glycoproteins listed in the SWISS-PROT database by the end of
1998
Glycoproteins with at
least one biochemically
characterized (`real')
glycosylation site
Glycoproteins with
at least one real
N-glycosylation site
and at least one real
O-glycosylation site
Glycoproteins with
at least one real
N-glycosylation site
and no real
O-glycosylation site
Glycoproteins with
at least one real
O-glycosylation site
and no real
N-glycosylation site
Sites
Entries
Sites
Entries
Sites
Entries
Sites
Entries
Potential N-glycosylation
sites (sequons)
2 066
697
289
80
1 679
582
98
35
Real glycosylation sites
1 965
749
556
80
1 041
582
368
87
Real N-glycosylation sites
1 279
662
238
80
1 041
582
0
0
Real O-glycosylation sites
686
167
318
80
0
0
368
87
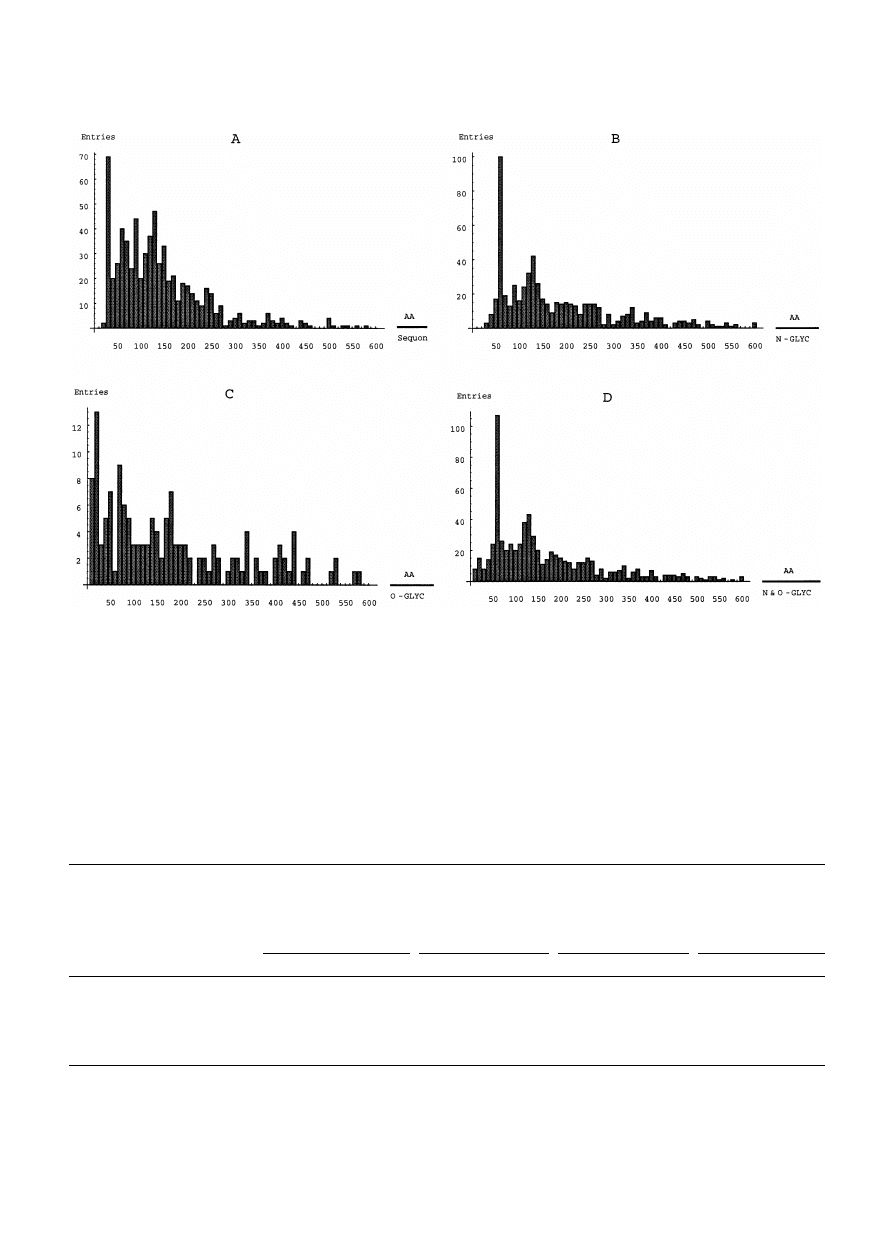
Fig. 2. Amino acid residues per sequon in all well characterized glycoproteins (A) and per real glycosylation site in the N-, O- and
N-,O-glycoproteins of the same group (B, C and D, respectively).
BBAGEN 24913 16-11-99
R. Apweiler et al. / Biochimica et Biophysica Acta 1473 (1999) 4^8
6
the present study, we denote as `real' glycosylation
sites only those sites where the 6 description s tag
does not contain any of the above strings. We con-
sider the glycosylation annotation of a SWISS-
PROT entry to be based on biochemical character-
ization if it contains at least one `real' (or occupied)
glycosylation site.
It should be noted that at present, at least one
third of all proteins in SWISS-PROT lack any bio-
chemical characterization and 12 921 are hypothetical
proteins, not yet proven to exist.
Furthermore, the proportion of biochemically
characterized proteins is getting smaller due to the
increasing number of predicted proteins derived
mainly from the fully sequenced genomes of di¡erent
organisms.
Of the 7942 listed glycoproteins, 1295 (16%) are
devoid of N-glycosylation sites, while 6647 (84%)
are N- (or N-,O-)glycoproteins. The latter contain a
total of 33 550 potential N-glycosylation sites, on
average 4.6 per molecule, which is more than that
found for all proteins containing such sites. Only a
small fraction of these, namely 749, have been thor-
oughly characterized with respect to their glycosyla-
tion patterns and complete information is available
as to the number and types of carbohydrate units per
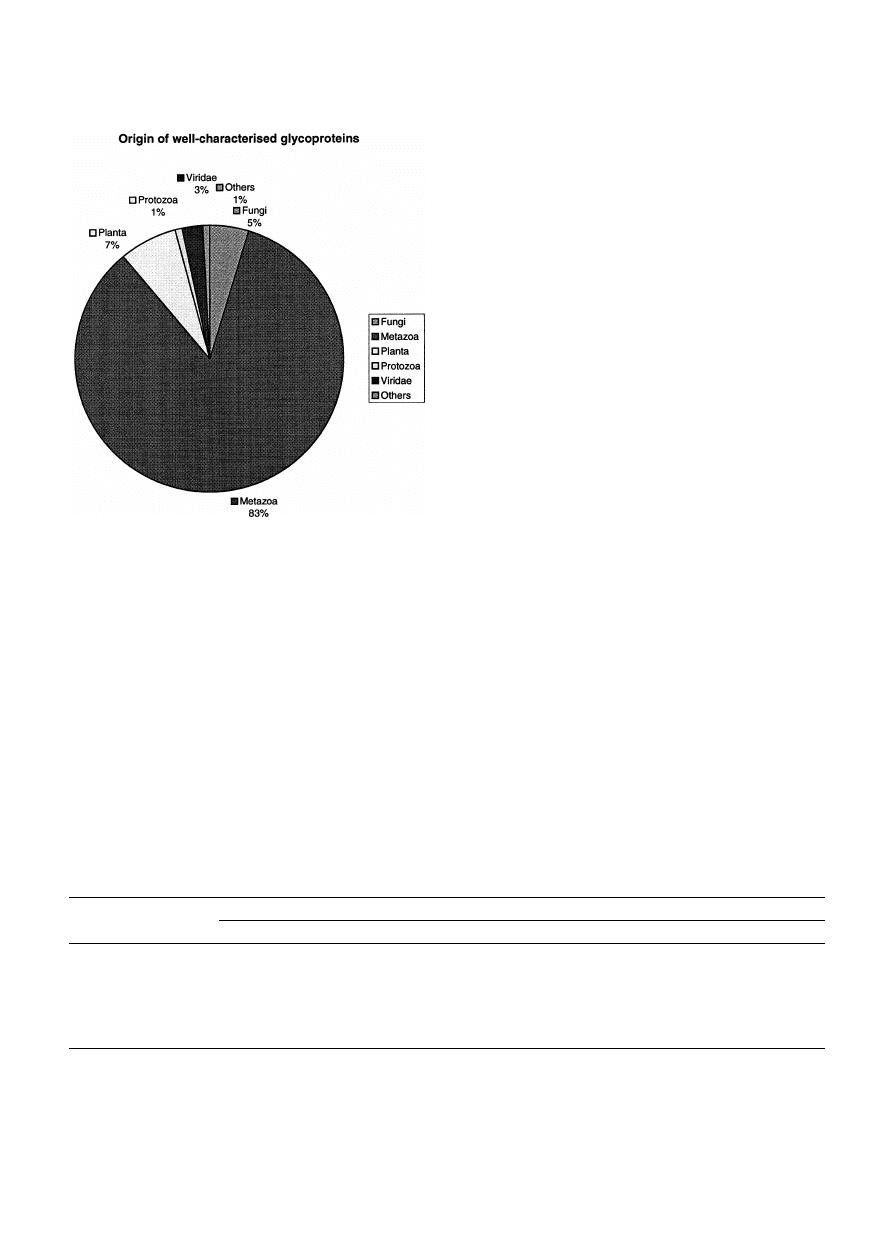
molecule (Table 2 and Fig. 1). The majority of these
glycoproteins (83%) is from animals, while the pro-
portion of those isolated from other sources is rela-
tively small, 12% from plants, including fungi, 3%
from viruses and 2% from microorganisms (bacteria
and protozoa) (Fig. 2).
Of the 749 well characterized glycoproteins, 662
are N- or N-,O-linked, containing in total 1968 se-
quons. This is an average of three potential N-glyco-
sylation sites per molecule of sequon containing pro-
tein, which is slightly lower than found for all sequon
containing proteins (Table 1). Of the sequons in the
well characterized glycoproteins, on average 1.9 per
molecule (i.e. about 2/3) are occupied. Typically, one
or two N-glycosidic carbohydrates are found in a
glycoprotein (Fig. 1B,D). They average 1.8 U per
N-linked glycoprotein and 3.0 U per N-,O-linked gly-
coprotein (Table 2), but their number may be as high
as 24 per molecule, as in gp120 of HIV [6]. The
number of O-linked units in the glycoproteins is
larger (Fig. 1C,D), averaging four per glycoprotein
(Table 2), with as many as 96 carbohydrate units
found in polysialoglycoproteins of Salmon ¢sh eggs
Fig. 3. Origin of the 749 well characterized glycoproteins in
SWISS-PROT.
Table 3
Spacing of sequons and real glycosylation sites in the 749 well characterized glycoproteins
Amino acid per
Sequon
Real N-site
Real O-site
Real N+O-site
a
Minimum
18.00
23.11
3.10
3.10
Maximum
1 669.00
3 412.00
2 813.00
3 412.00
Mean
159.04
249.92
269.90
231.66
Median
121
144
167
133
Range
1 651.00
3 388.89
2 809.90
3 408.90
S.D.
164.07
348.09
338.83
335.80
a
In N-,O-glycoproteins.
BBAGEN 24913 16-11-99
R. Apweiler et al. / Biochimica et Biophysica Acta 1473 (1999) 4^8
7

[7]. The range of spacing of the sequons and the real
glycosylation sites is extremely wide (Fig. 3 and Ta-
ble 3).
Assuming that the well characterized glycoproteins
are a representative sample of the glycoproteins
present in nature, some three quarters of all glyco-
proteins should be N-linked, about one tenth N-,O-
linked and about one eighth just O-linked. Based on
the rate of occupancy of the sequons in the well
characterized glycoproteins, the 48 636 sequon con-
taining proteins of SWISS-PROT should each carry
on average two N-linked carbohydrates. This is close
to the average number of such units per molecule of
well characterized glycoprotein. It is thus highly
likely that most of the sequon containing proteins
will prove to be glycosylated. To this estimate should
be added the 10% proteins expected to be O-glyco-
sylated. We conclude therefore that more than half
of all proteins in nature will eventually be found to
be glycoproteins.
References
[1] N. Sharon, Complex Carbohydrates, their Chemistry, Bio-
synthesis and Functions. Addison Wesley, Reading, MA,
1975, pp. 33^35.
[2] C.G. Gahmberg, M. Tolvanen, Why mammalian cell surface
proteins are glycoproteins, Trends Biochem. Sci. 21 (1996)
308^311.
[3] J. Montreuil, The history of glycoprotein research, a person-
al view, in: J. Montreuil, J.F.G. Vliegenthart and H.
Schachter (Eds.), Glycoproteins. Elsevier, Amsterdam,
1995, p. 1.
[4] N. Sharon and H. Lis, Glycoproteins: structure and func-
tion, in: H.-J. Gabius and S. Gabius (Eds.), Glycosciences-
Status and Perspectives. Chapman and Hall, Weinheim, Ger-
many, 1997, p. 133.
[5] A. Bairoch, R. Apweiler, The SWISS-PROT protein se-
quence data bank and its supplement TrEMBL in 1999,
Nucleic Acid Res. 27 (1999) 49^54.
[6] C.K. Leonard, M.W. Spellman, L. Riddle, R.J. Harris, J.N.
Thomas, T.J. Gregory, Assignment of intrachain disul¢de
bonds and characterization of potential glycosylation sites
of the type 1 recombinant human de¢ciency virus envelope
glycoprotein (gp120) expressed in Chinese hamster ovary
cells, J. Biol. Chem. 265 (1990) 10373^10382.
[7] J.K. Kitajima, Y. Inoue, S. Inoue, Polysialoglycoprotein of
Salmonidae ¢sh eggs. Complete structure of 200-kDa poly-
sialoglycoprotein from the unfertilized eggs of rainbow trout
(Salmo gairdneri), J. Biol. Chem. 261 (1986) 5262^5269.
BBAGEN 24913 16-11-99
R. Apweiler et al. / Biochimica et Biophysica Acta 1473 (1999) 4^8
8
Wyszukiwarka
Podobne podstrony:
Effecto of glycosylation on the stability of protein pharmaceuticals
Thomas Aquinas And Giles Of Rome On The Existence Of God As Self Evident (Gossiaux)
On The Specification Of Moving Coil Drivers For Low Frequency Horn Loaded Loudspeakers (W Marshall L
1The effects of hybridization on the abundance of parental taxa depends on their relative frequency
Interruption of the blood supply of femoral head an experimental study on the pathogenesis of Legg C
Ogden T A new reading on the origins of object relations (2002)
Newell, Shanks On the Role of Recognition in Decision Making
On The Manipulation of Money and Credit
Dispute settlement understanding on the use of BOTO
Fly On The Wings Of Love
31 411 423 Effect of EAF and ESR Technologies on the Yield of Alloying Elements
Crowley A Lecture on the Philosophy of Magick
On the Atrophy of Moral Reasoni Nieznany
Effect of magnetic field on the performance of new refrigerant mixtures
94 1363 1372 On the Application of Hot Work Tool Steels for Mandrel Bars
76 1075 1088 The Effect of a Nitride Layer on the Texturability of Steels for Plastic Moulds
Gildas Sapiens On The Ruin of Britain
więcej podobnych podstron