
Translacja:
Translacja:
elongacja
elongacja

Wobble Base Pairing
The presence of more than one codon for a
given amino acid raises the issue of how many
tRNAs are necessary to read the code.
You could have one for each codon.
It turns out that there are less than this.
The reason has to do with what Crick called
"wobble base pairing".
Wobble refers to non Watson-Crick base
pairing that can take place at the third position
of the codon. Here is figure that shows Watson-
Crick versus wobble pairing:
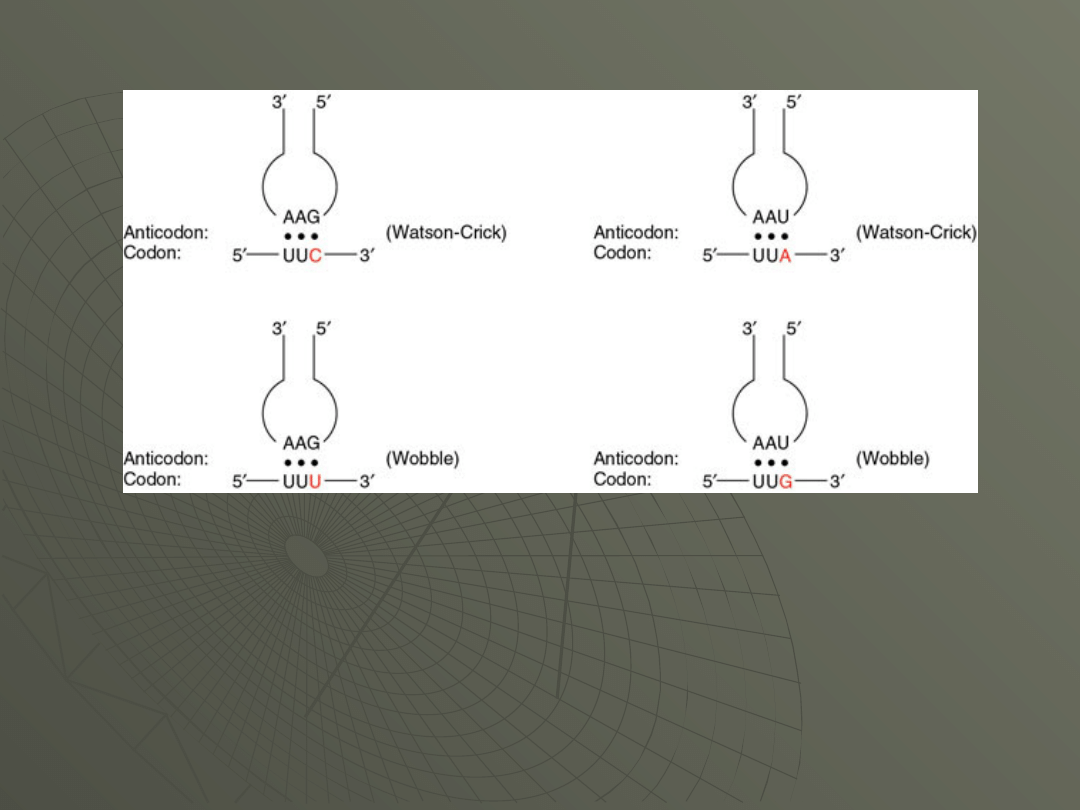
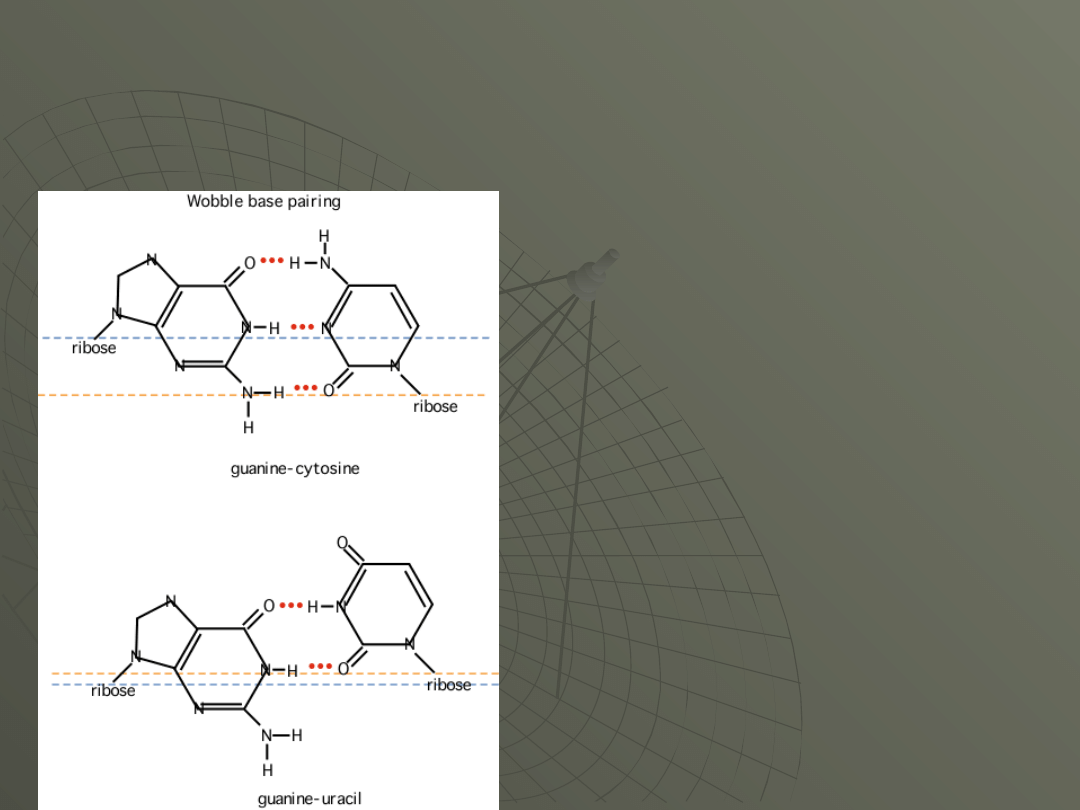
The wobble pair is so-called because the base
has shifted ("wobbled") in order to make the
hydrogen bonding work. Here's how it looks:
For the top Watson-
Crick pair (GC) the
dotted lines show the
relative positions of the
ribose sugars.
In the bottom wobble
pair (GU) the dotted
lines show how the
sugars are in a
different relative
position.
The effect of wobble is
to require fewer tRNAs
to translate the code.
Wobble Rules
5' position in
anticodon
3' position in codon
G
pairs
with
C or U
C
pairs
with
G
A
pairs
with
U
U
pairs
with
A or G
I
pairs
with
A, U, or C
The anticodon position in the tRNA can also have the
base inosine (I), a purine that is not present in the
messenger RNA (codon).
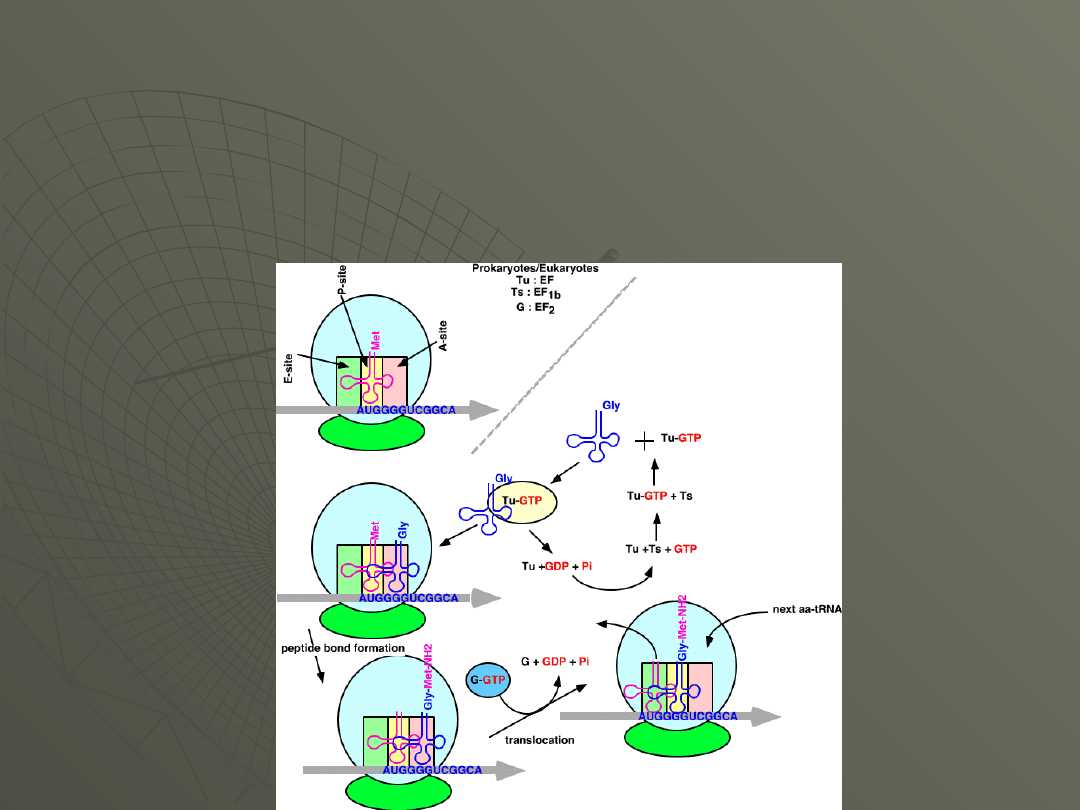
Elongation
The events of elongation are essentially the
same in both prokaryotes and eukaryotes. Here
is a figure that shows these events in
summary:

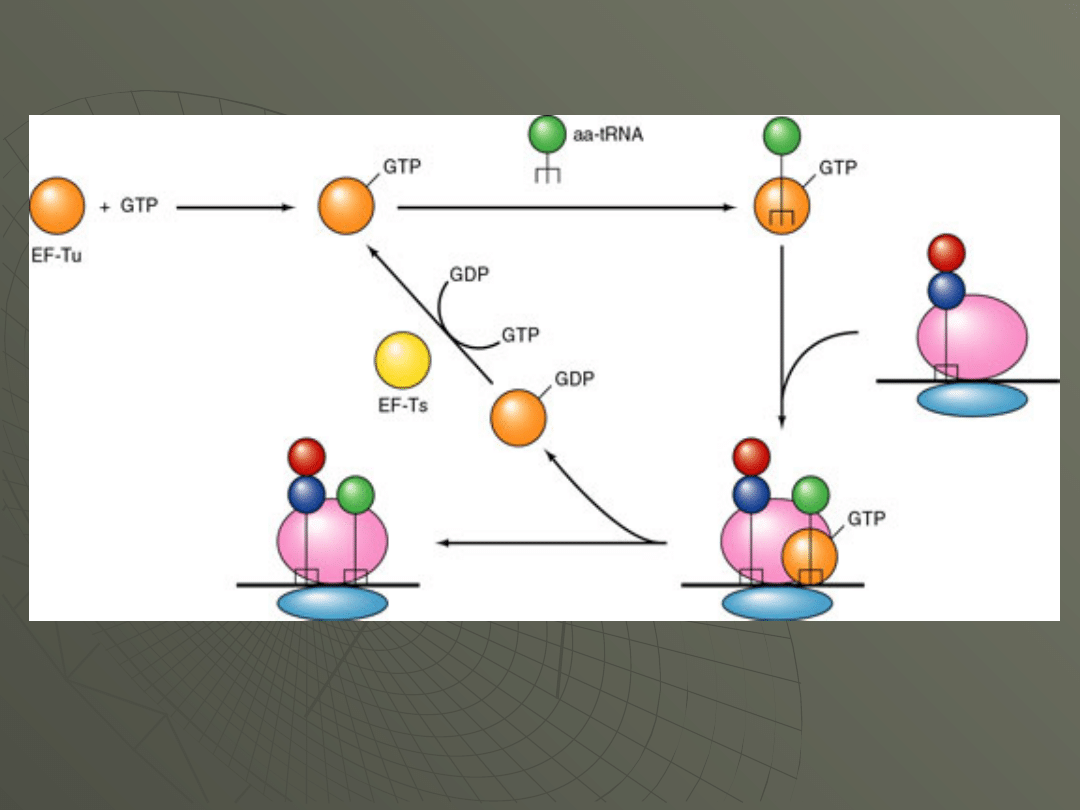
Step 1: Entry (binding) of the next
aminoacyl-tRNA to the A site.
The next amino acid to be added to the protein
chain is brought by its tRNA to the A site of the
ribosome.
This is carried out by EF-Tu (EF in eukaryotes).
This requires that Tu be bound to a GTP.
The placement of the aminoacyl-tRNA into the A
site is at the expense of energy, and the GTP is
hydrolyzed to GDP which leaves the protein.
At this point the EF-Tu must be recycled by the
addition of another GTP. This is accomplished by
EF-Ts (EF1b in eukaryotes).

Step 2: Formation of the Peptide Bond
The tRNA that was already present in the
ribosome (say, the initiator tRNA) is in the P
site of the ribosome.
A bond is now formed between the COOH
terminus of the amino acid or peptide chain on
the tRNA in the P-site (actually where it's
linked to the tRNA) and the NH
2
terminus of
the amino acid in the A-site.
The reaction is catalyzed by a part of the large
subunit of the ribosome.
The actual catalytic activity is probably a
function of the large rRNA and is therefore a
ribozyme reaction.
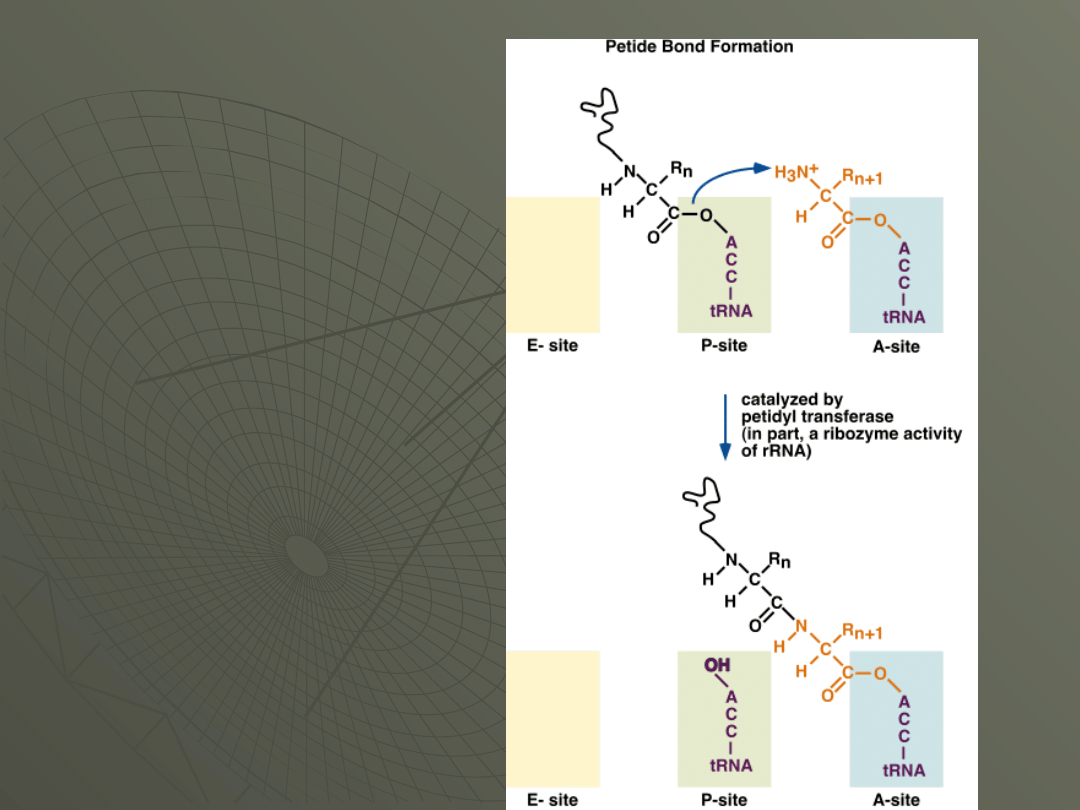
Here is the formation
of the peptide bond:
The end of this
reaction, the
growing peptide
chain is carried by
the tRNA in the A
site of the ribosome.

Step 3: Translocation
The ribosome is now going to move relative to
the messenger RNA, down the mRNA in the 5'
to 3' direction.
The net result of this movement will be to
transfer the tRNA carrying the polypeptide into
the P-site, and the empty tRNA into the E-site,
leaving the A-site vacant, positioned over the
next codon and ready for the next aminoacyl-
tRNA.
This reaction requires another elongation
factor, EF-G (EF2 in eukaryotes).
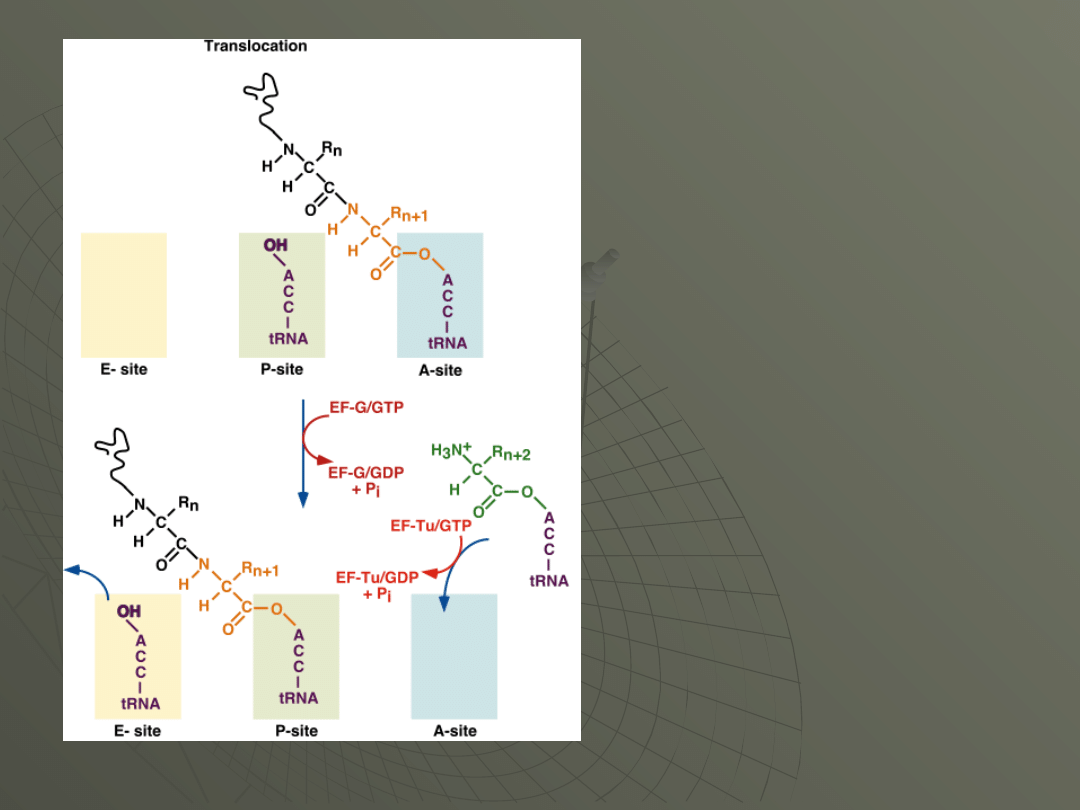
Once the empty
tRNA is in the E-
site, it leaves the
ribosome and the
entire machine is
ready for the next
elongation step.
This means
bringing the next
aminoacyl-tRNA
into the A-site,
again using EF-
Tu/GTP.

The Open Reading Frame
Elongation continues until all of the amino
acids that make up the protein have been
added to the chain.
The protein is defined by the open reading
frame (ORF).
An open reading frame is the sequence of
codons from (and including) the start codon
(usually AUG) to the stop codon (UAG, UAA,
or UGA).
Document Outline
- Slide 1
- Slide 2
- Slide 3
- Slide 4
- Slide 5
- Slide 6
- Slide 7
- Slide 8
- Slide 9
- Slide 10
- Slide 11
- Slide 12
- Slide 13
Wyszukiwarka
Podobne podstrony:
2014 BPEG część 6 elongacja translacji
33 Przebieg i regulacja procesu translacji
BM7 Translacja
Ekspresja informacji genetycznej-transkrypcja i translacja, NAUKA
In literary studies literary translation is a term of two meanings rev ag
Early Theories of Translation
Erratum to Revised Transliminality scale
historical identity of translation
dokumenty-translation-all
Translatoryka bibliografia
Przeklad ogolny- zaliczenie, Filologia angielska, Notatki I rok, general translation
translantologia - odpowiedzi(1), SPECJALIZACJA pielęgniarstwo operacyjne, Testy
Choosing a Bible Understanding Bible Translation Differences
translacja
11 Translatorisches Handeln
2014 BPEG część 8 regulacja translacji
więcej podobnych podstron