
TRANSLACJA
TRANSLACJA

[MCB 411/411H
Module 17
The Mechanism of Translation]
An overview of translation
In order for proteins to be made in cells, the
following components are necessary:
-messenger RNA (mRNA)
-ribosomes (complexes of protein and
ribosomal RNA [rRNA])
-transfer RNA (tRNA)

-various protein factors
-amino acids
-energy (ATP and GTP)

The process of translation has four
stages:
1.tRNA charging (adding the correct amino
acid to the correct tRNA)
2. initiation (assembly mRNA, ribosomes, and
tRNA)
3. elongation (reading the code and
converting the information into a peptide)
4. termination (ending the synthesis of a
protein)
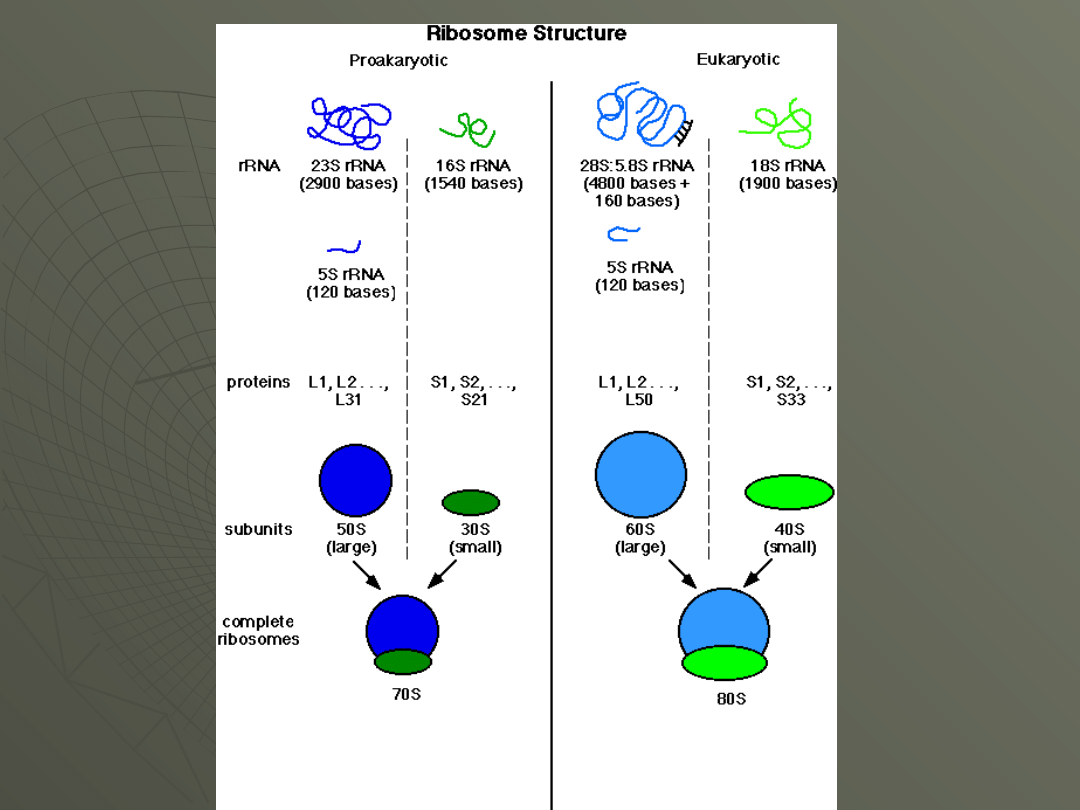
Transfer RNA
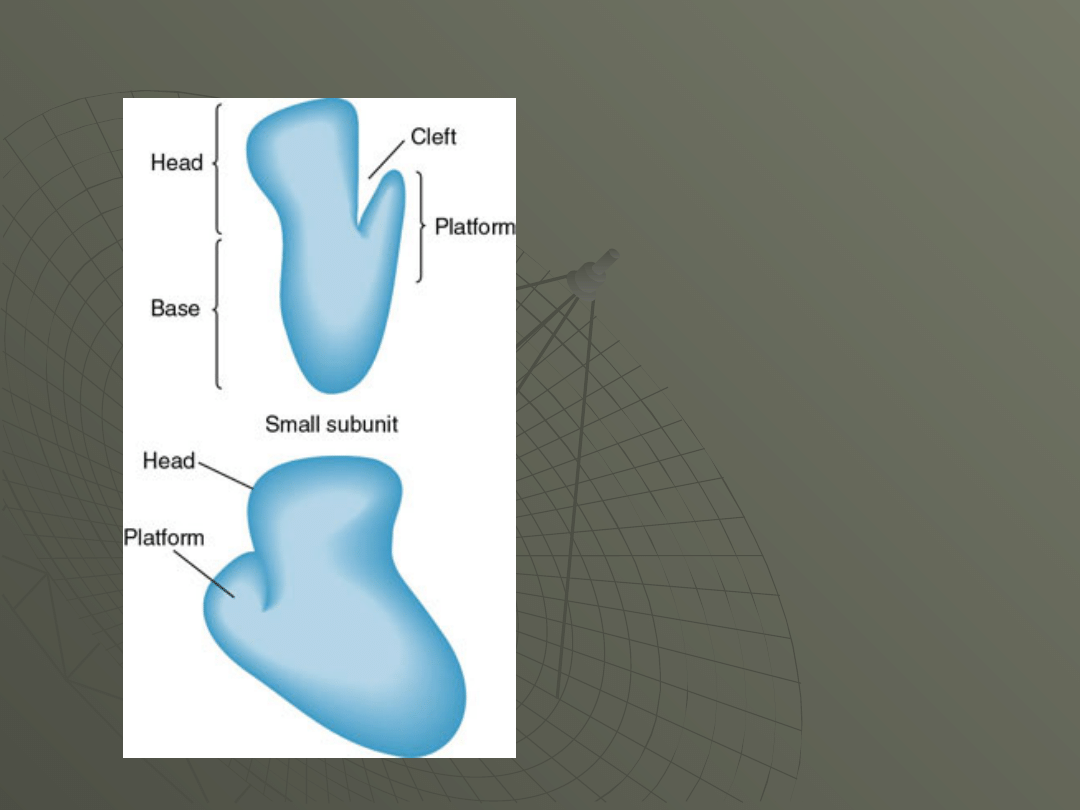
E. coli 30S subunit
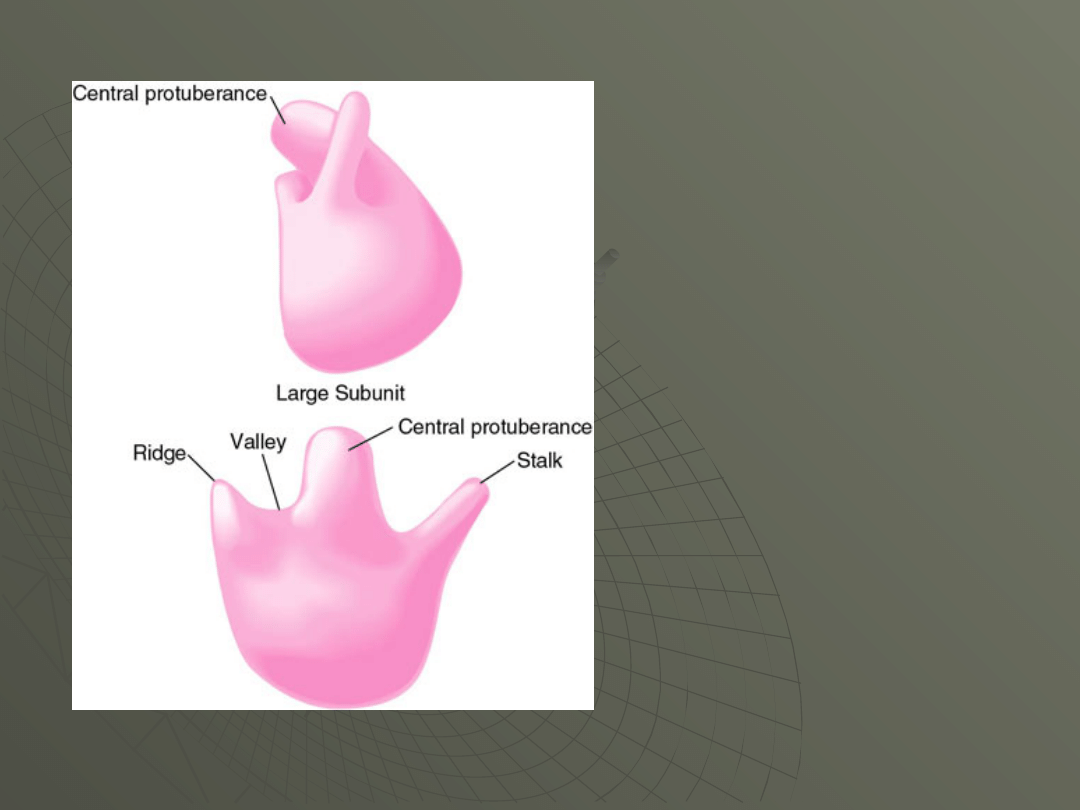
E. coli 50S subunit
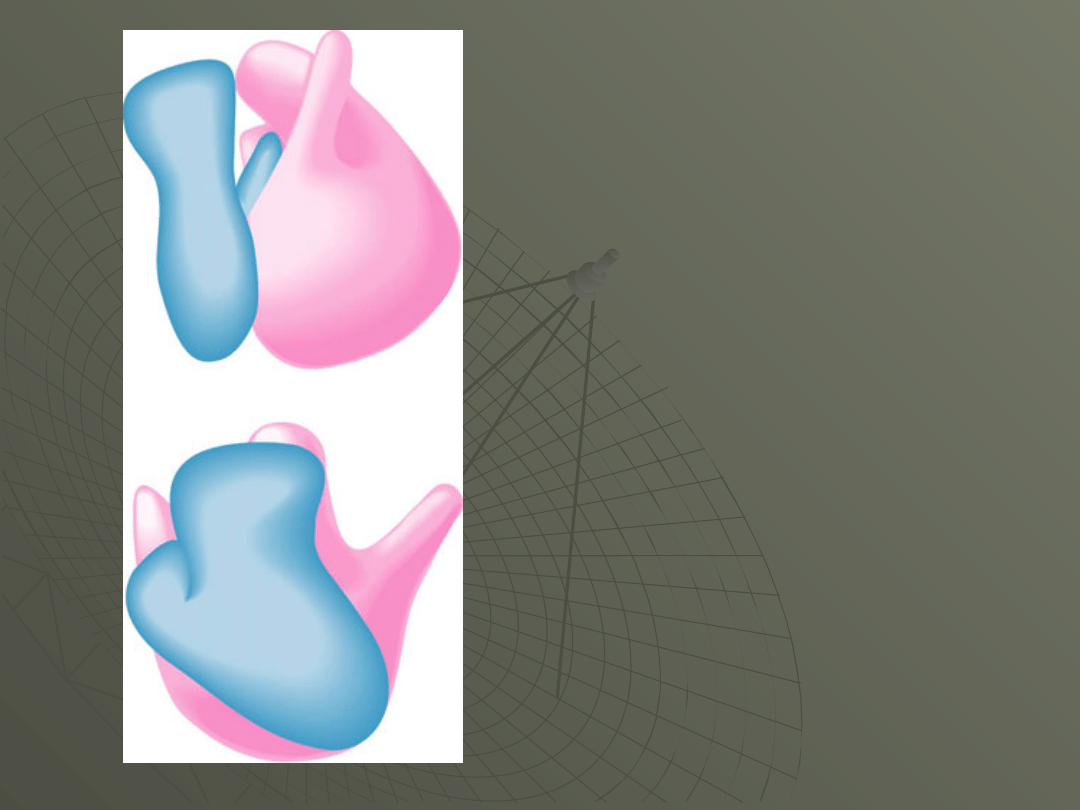
E. coli 70S ribosome
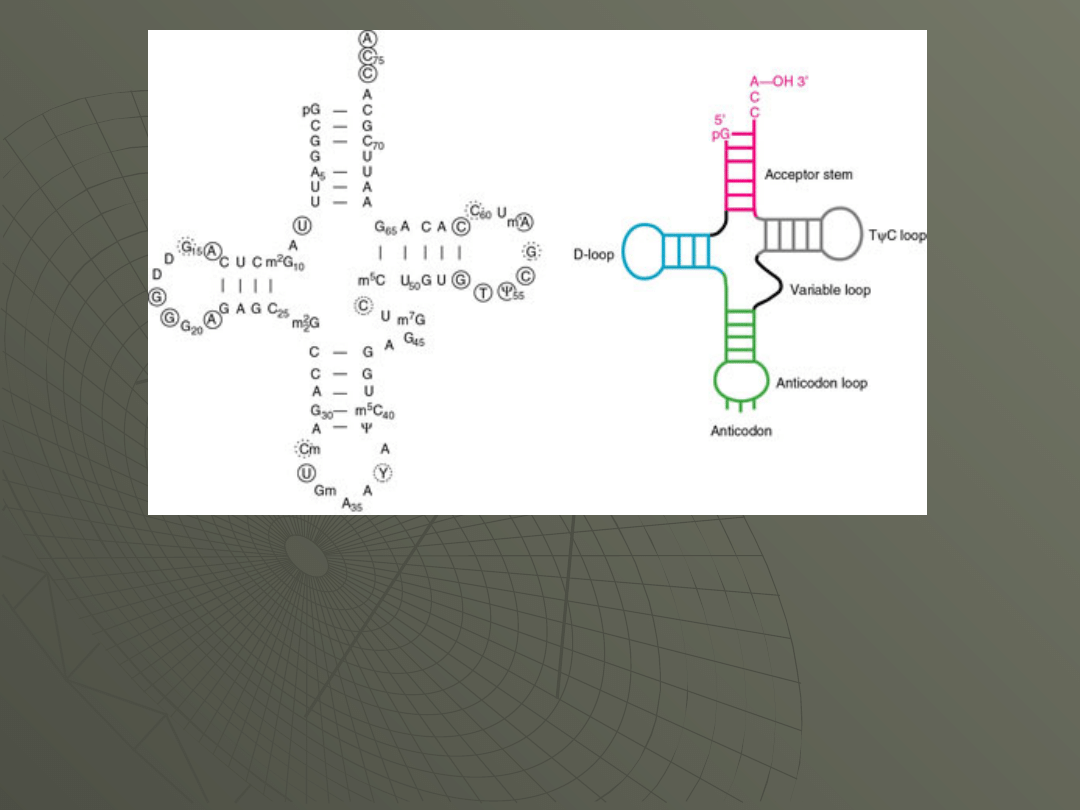
The general structure shown on the left has the
important features that you will find in all tRNAs.
This includes the base paired stems, including the
acceptor stem, and the loops (the D-loop, the TYC-
loop and the anticodon loop). D and Y are symbols
that indicate modified bases that are present in
tRNAs and not generally found in other RNAs.
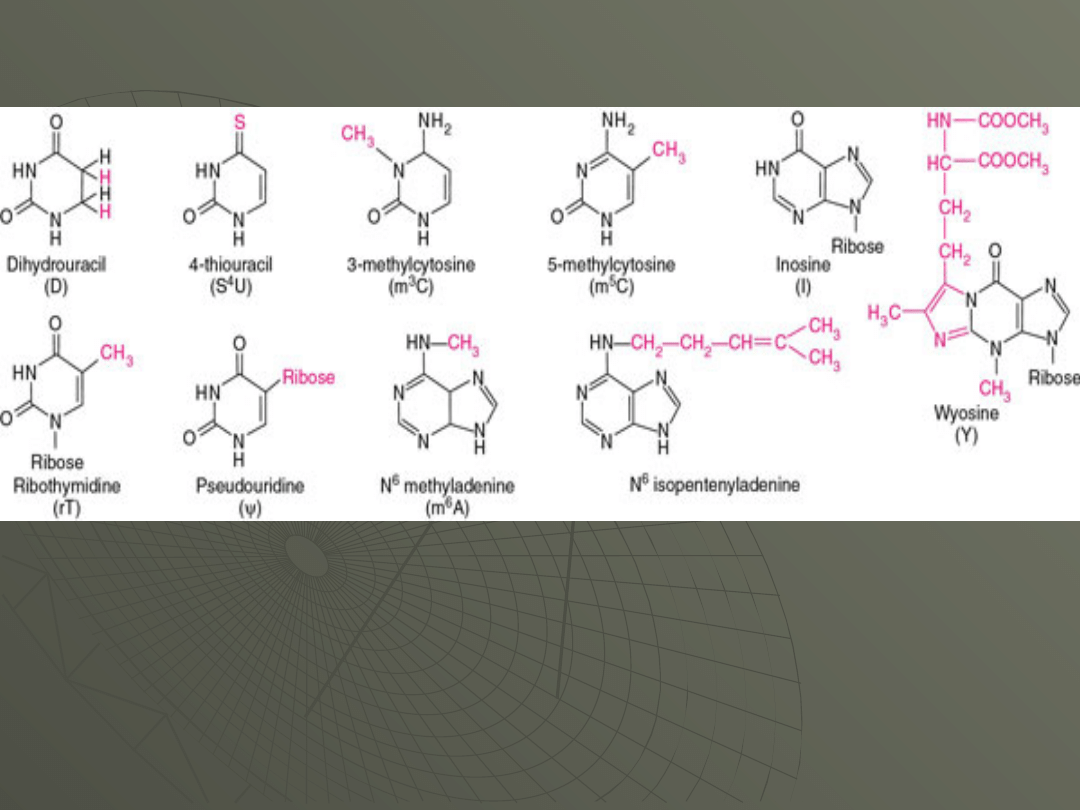
These modified bases are shown in Figure:
The significance of the modified bases is not
totally clear. The modifications take place after
the transcription of the tRNA from the DNA
template. Data suggest that if the bases are left
unmodified the tRNA does not function properly.
What their exact role is remains unknown.
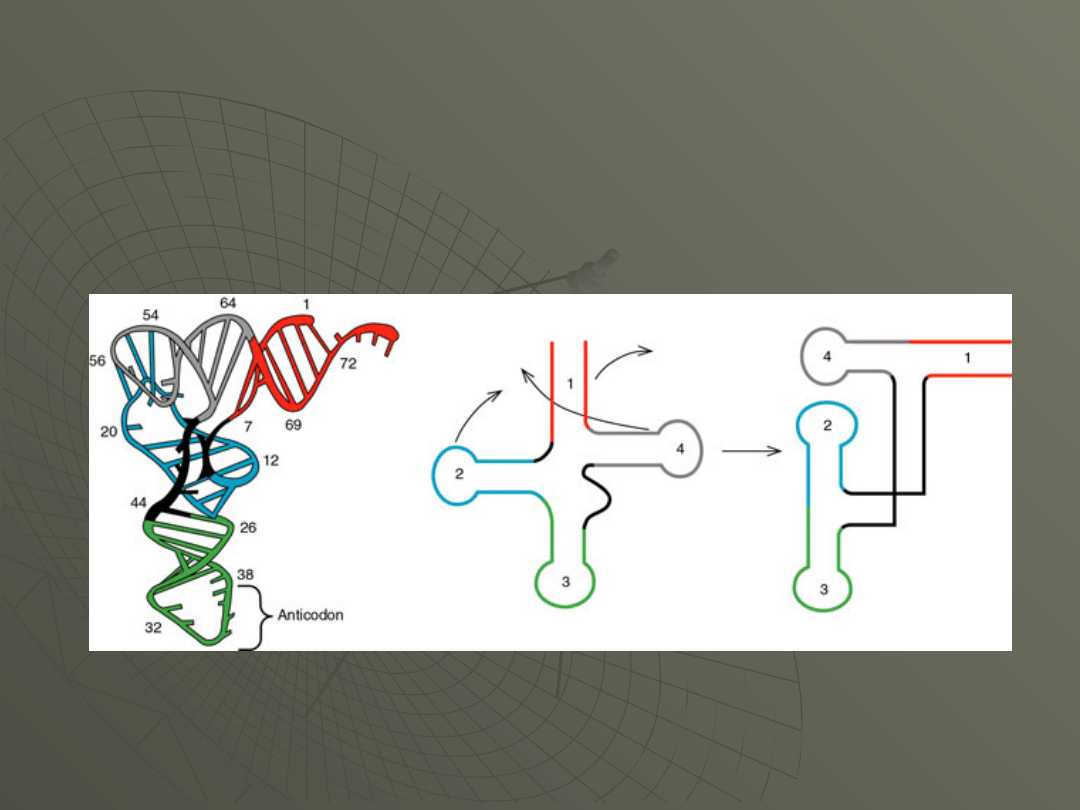
Of course the cloverleaf structure is just a cartoon representation.
Another representation of the molecule comes from X-ray
crystallography. In this case, it appears that the molecule is folded up
into the form shown in Figure

The various regions of the molecule are color
coded as in Figure.
This folded structure is thought to be more
representative of the molecule as it might
occur within the cell. These models of tRNAs
are quite striking in three dimensional
representation:
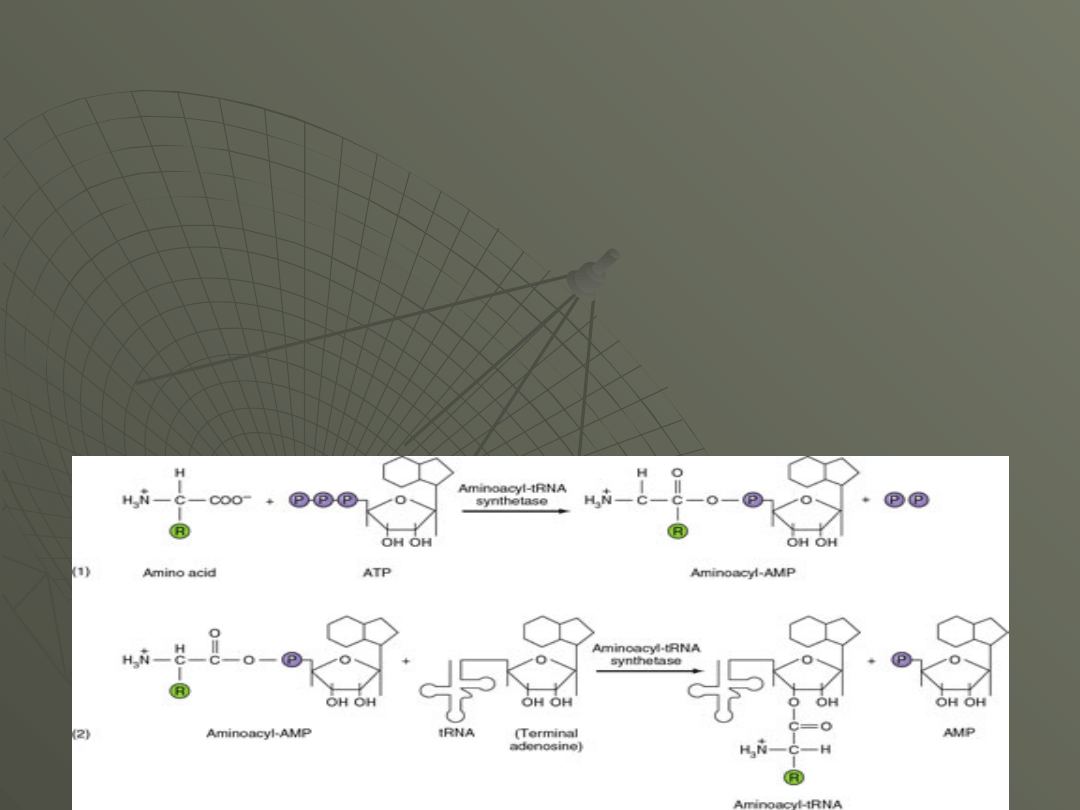
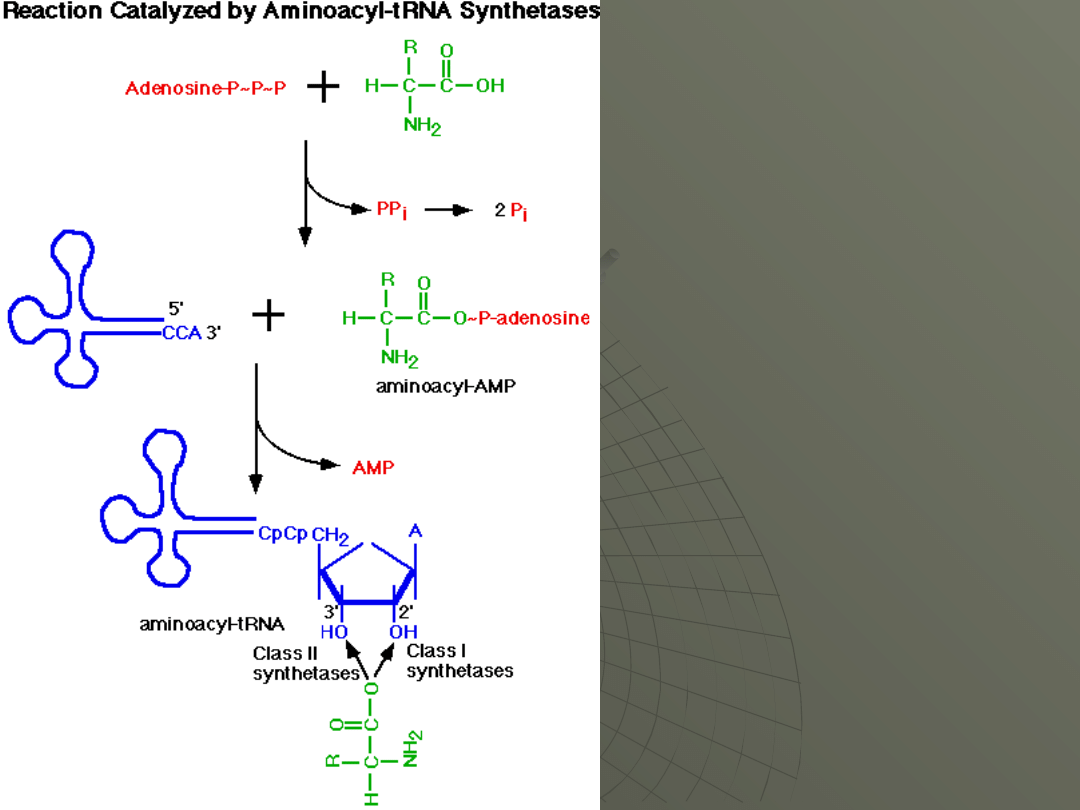
Transfer RNA Charging
The process of adding the correct amino acid to
the tRNA responsible for bringing to the
ribosome is called "charging." The word
"charge" here is used in the sense of "loading,"
as in charging a battery.
The charging reaction is catalyzed by a set of
enzymes called aminoacyl tRNA synthetases.

The enzyme catalyzes a two-step reaction:
1.In the first step, utilizes ATP and the amino
acid is energized by the covalent addition of
AMP, joining the two molecules by making a
bond between the phosphate of AMP and the
COOH of the amino acid.
This splits of the pyrophosphate of ATP,
which is immediate broken down into to
inorganic
phosphates
by
the
enzyme
pyrophosphatase.
Therefore, the energy of two phosphodiester
bonds drives this reaction.

2. The second step is the transfer of the
amino acid to the 3' or 2' OH of the terminal A
of the tRNA.
The product of the reaction is an aminoacyl-
tRNA and AMP.
The overall reaction is as follows:
amino acid (aa) + ATP + tRNA --> aminoacyl-
tRNA + AMP + 2 Pi
The link to the tRNA can be initially at either
the 3' OH or the 2' OH.
The 3' end of every tRNA is the sequence
---CCAOH.
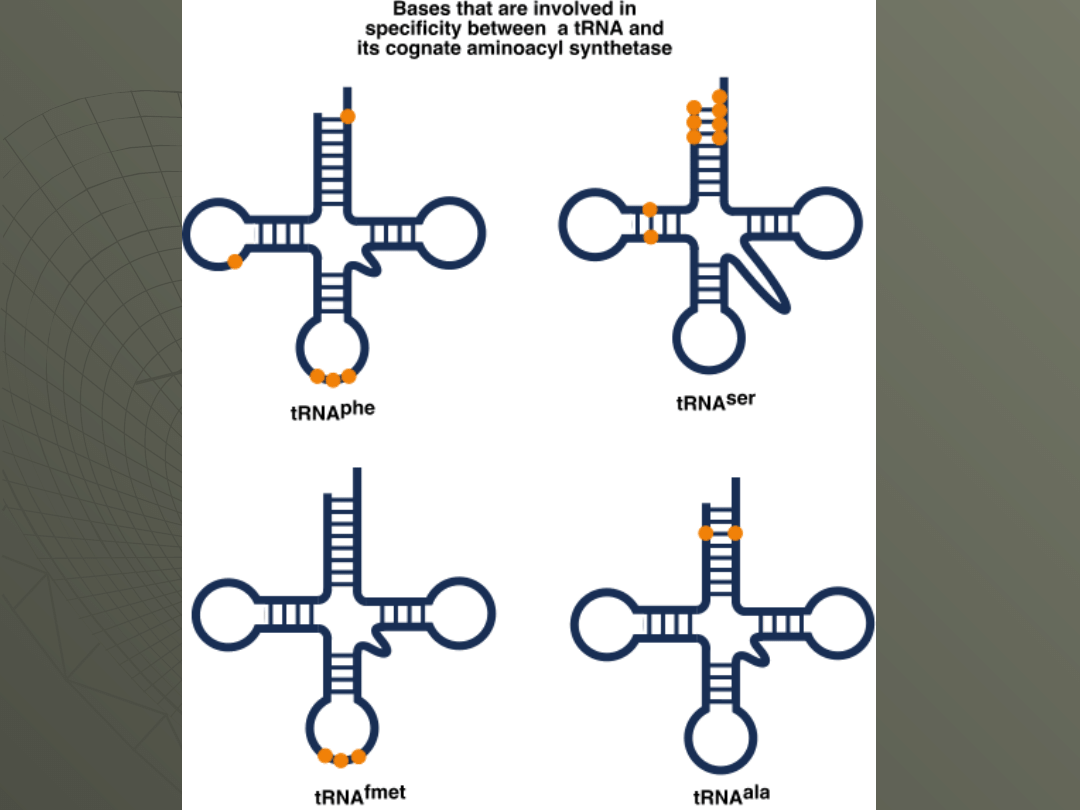
The specificity of
these enzymes
(aminoacyl tRNA
synthetases) is
determined by the
structure of the
tRNA.
Although you would
think that the
anticodon provides
all of the information,
an analysis of various
tRNA structures
shows that the
important bases
might be at other
locations in the
molecule.

Prokaryotic Initiation
Protein synthesis in all cells begins (is initiated)
by the association of mRNA, tRNA and
ribosomes.
This initiation is mediated by a series of factors
appropriately called initiation factors (IFs).
In Prokaryotes there are three: IF-1, IF-2, and
IF-3.
The first step is to make sure that the ribosome
is dissociated into it's subunits.
This is accomplished by two of these factors.
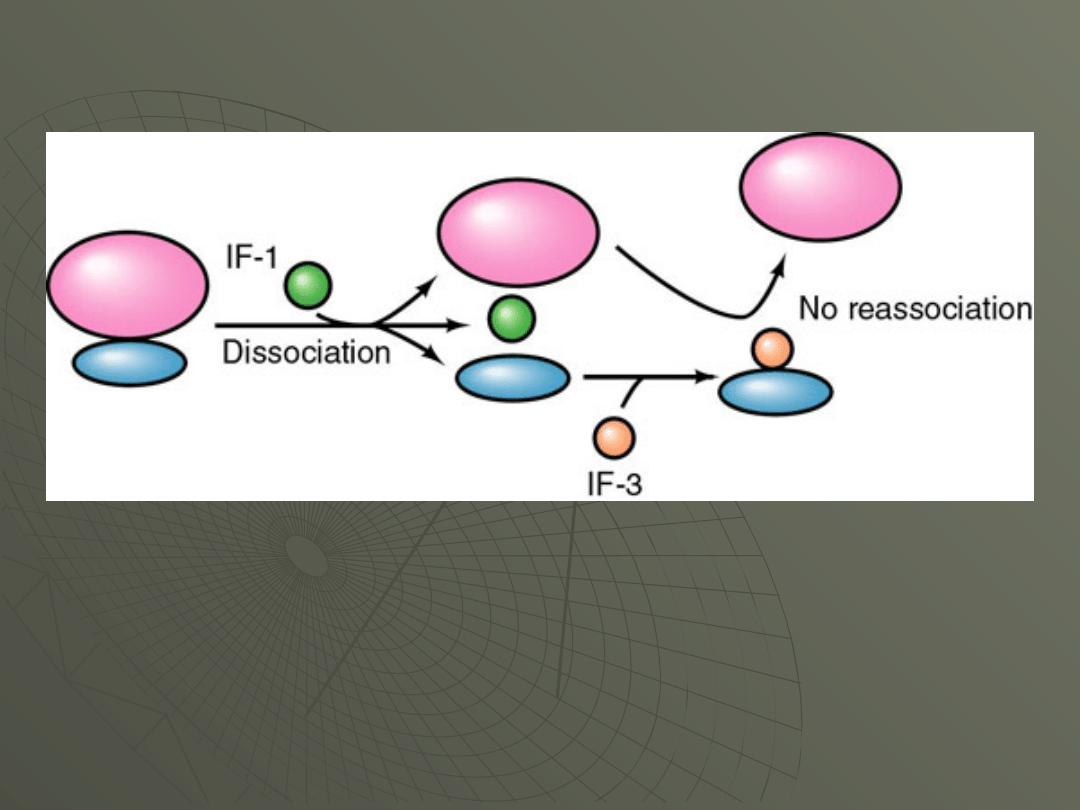
This sequence of events is diagrammed in Figure:
In the cell intact ribosomes (70S ribosomes)
interact with IF-1 to cause dissociation into the
two subunits, 30S (the small subunit) and 50S
(the large subunit). IF-3 then binds to the 30S
subunit and keeps the two from reassociating
until the time is correct.

The first event of interaction between the
components of translation is the formation of
the 30S initiation complex. This involves the
30S subunit, the mRNA, and a special initiator
tRNA.
The code word that says "start protein
synthesis" is AUG, the same code word that
means "methionine".
As you might expect, the tRNA that is the
initiator will be one that can read this code
word.
However, in prokaryotes, this special tRNA
carries a modified version of the amino acid
methionine.
Figure shows the structure of methionine and
N-formyl-methionine:
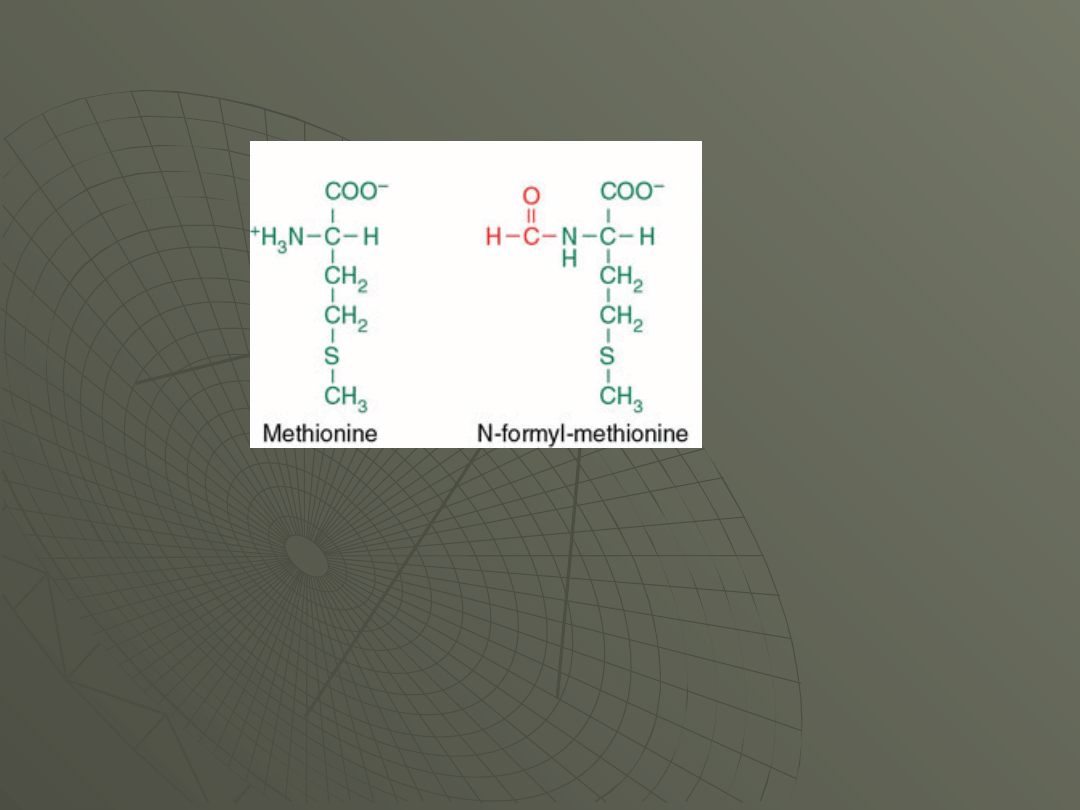
Figure shows the structure of methionine and
N-formyl-methionine:
In prokaryotes, the 30S subunit of the ribosome,
along with fMet-tRNAMetf, interacts directly with
the start codon (AUG) that begins the protein to
be translated. All three initiation factors (IF-1, IF-
2, IF-3) are involved in this process. IF-2 carries
out it's function in this event by having a GTP
bound to it. The energy from this GTP will be
used a bit later.

The precise placement of the 30S subunit at
the start codon is achieved by a base pairing
between the 3' end if the 16S rRNA of the 30S
subunit and sequences found just upstream
from the initial AUG.
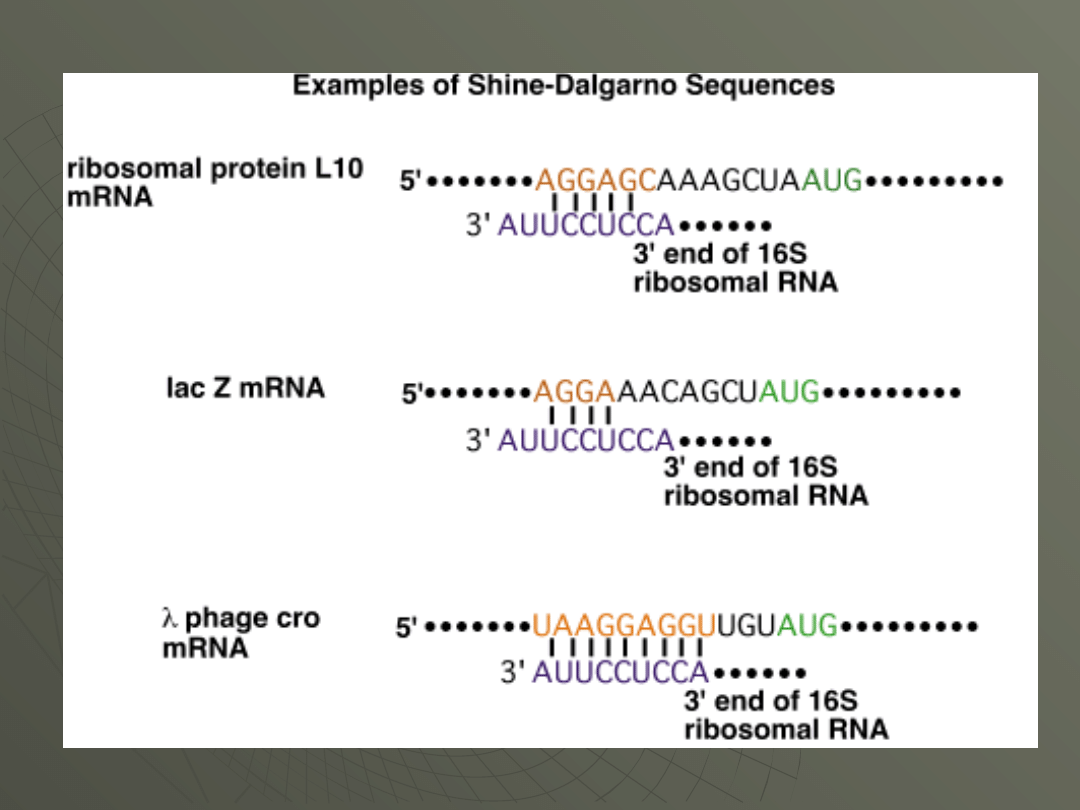
This base-pairing interaction was discovered
by John Shine and Lynn Dalgarno and the
sequence in the mRNA is therefore called the
Shine-Dalgarno sequence (SD sequence).
This base-pairing is shown in the following
figure:

Once the 30S subunit is located correctly, the
initiator tRNA is positioned over the AUG codon
that begins the protein.
This forms the 30S initiation complex.
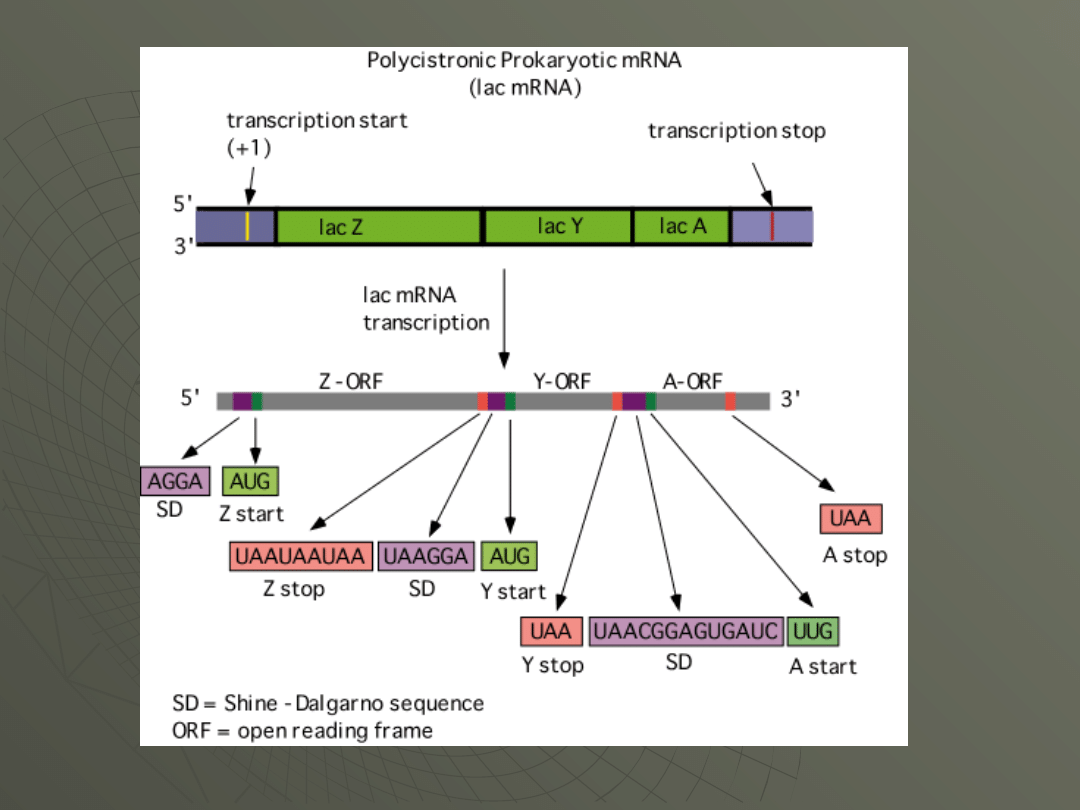
The SD-sequence explains how prokaryotic
mRNA can be polycistronic.
Since the 30S subunit finds its way to each
start codon, several different starts can be
made on the same mRNA, if they have the SD-
sequence upstream.
Here's the lac messenger RNA as an example,
showing the location of the SD-seqences
upstream of each gene:

Next, the 50S subunit joins this complex, to
form the 70S initiation complex.
The three factors now leave and the GTP that
is attached to IF-2 is hydrolyzed to GDP and
Pi.
All of the events of prokaryotic initiation are
summarized in the following table and in
Figure:
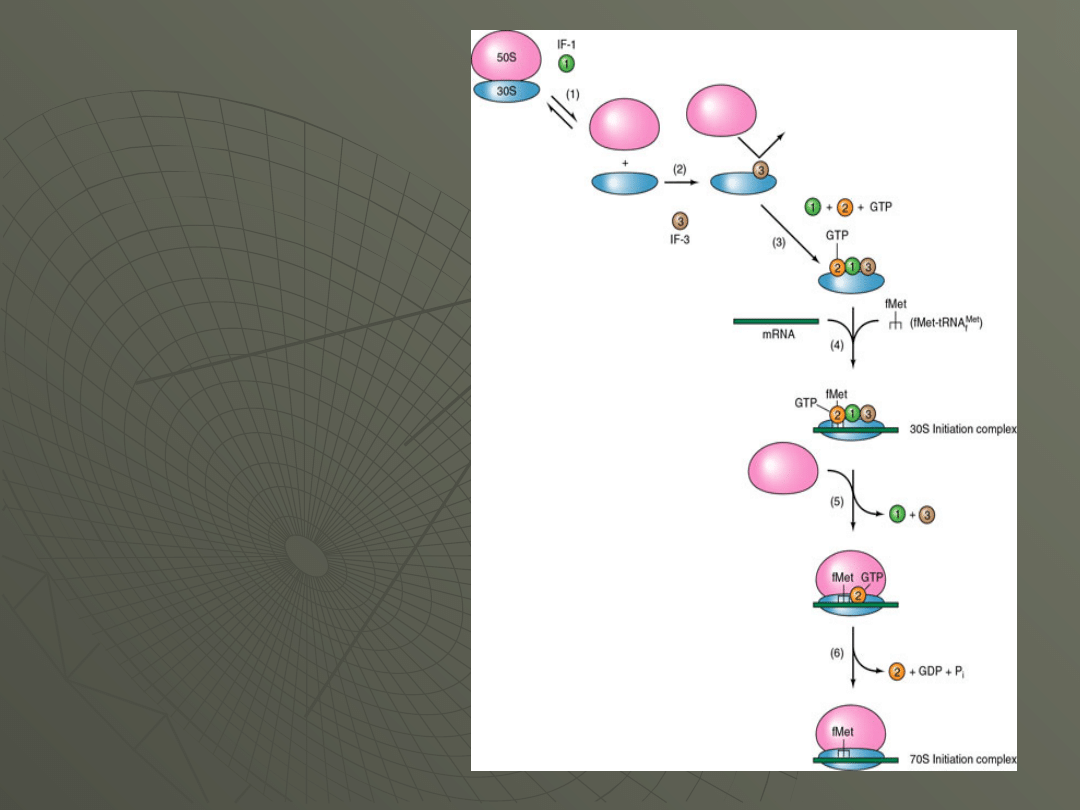
Steps in Formation of
70S initiation
Complex:
1. dissociation of the
70S ribosome into the
30S and 50S subunits
(IF-1)
2. binding of IF-3 to 30S
3. binding of IF-1 and
IF-2/GTP to 30S
4. joining of 30S/IF's
with initiator tRNA and
mRNA to form 30S
initiation complex
5. Binding of 50S to 30S
complex, with loss of IF-
1 and IF-3
6. dissociation of IF-2
with hydrolysis of GTP
to GDP and Pi

Eukaryotic Initiation
The beginning of protein synthesis in
eukaryotes also involves the association of
ribosome, tRNA, and mRNA.
However, unlike the prokaryotes, this
association takes place in a very different way.
Remember that the 5' end of eukaryotic
mRNAs has a methylated cap.
Marilyn Kozak discovered that eukaryotic
ribosomes do not bind directly to the region of
the messenger RNA that contains the AUG
initiation codon.
Instead, the 40S ribosomal subunit starts at
the 5' end and "scans" down the message until
it arrives at the AUG codon.
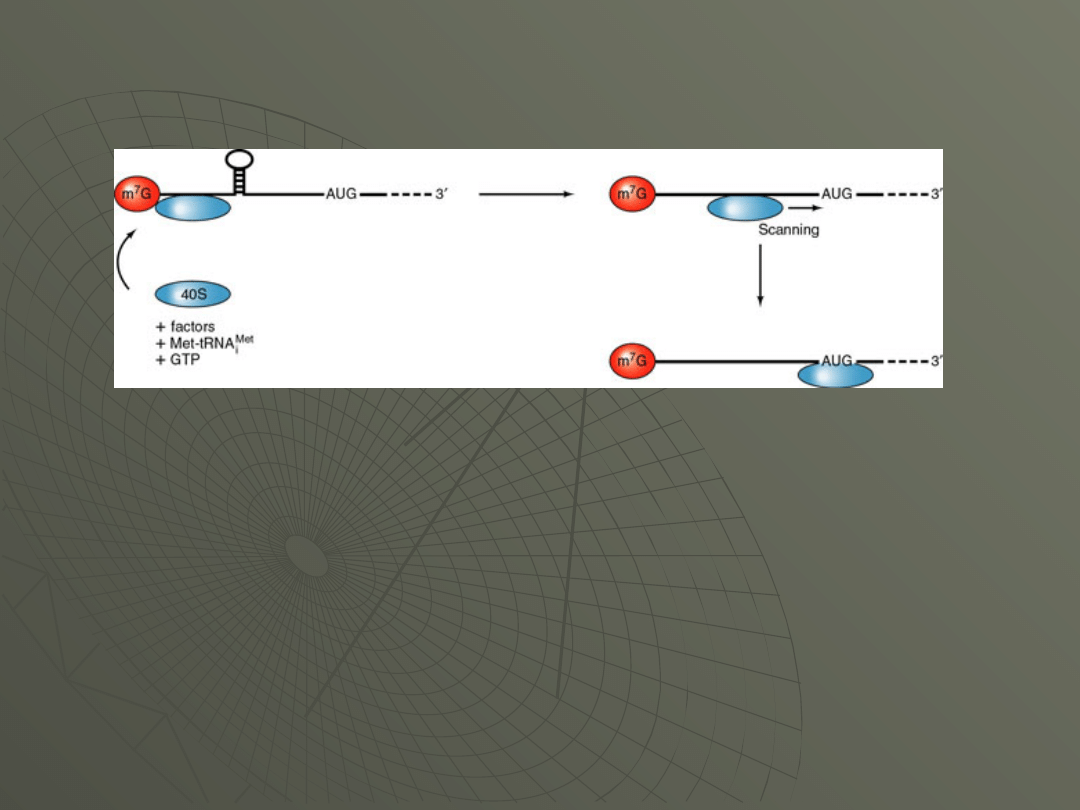
This is diagrammed in Figure:
The AUG that is recognized likely sits within
a certain sequence context with the
following consensus:
CCRCCAUGG
where R is a purine (A or G).

In addition to the 40S subunit, this requires
energy (GTP), eukaryotic initiation factors (eIFs,
see below) and an initiator tRNA.
Unlike the prokaryotic case, the methionine
carried by this tRNA is not formlyated. The tRNA
is called tRNAi.
When it carries methionine, it is called Met-
tRNAiMet.
The formation of the initiation complex in
eukaryotes proceeds as outline in Figure:
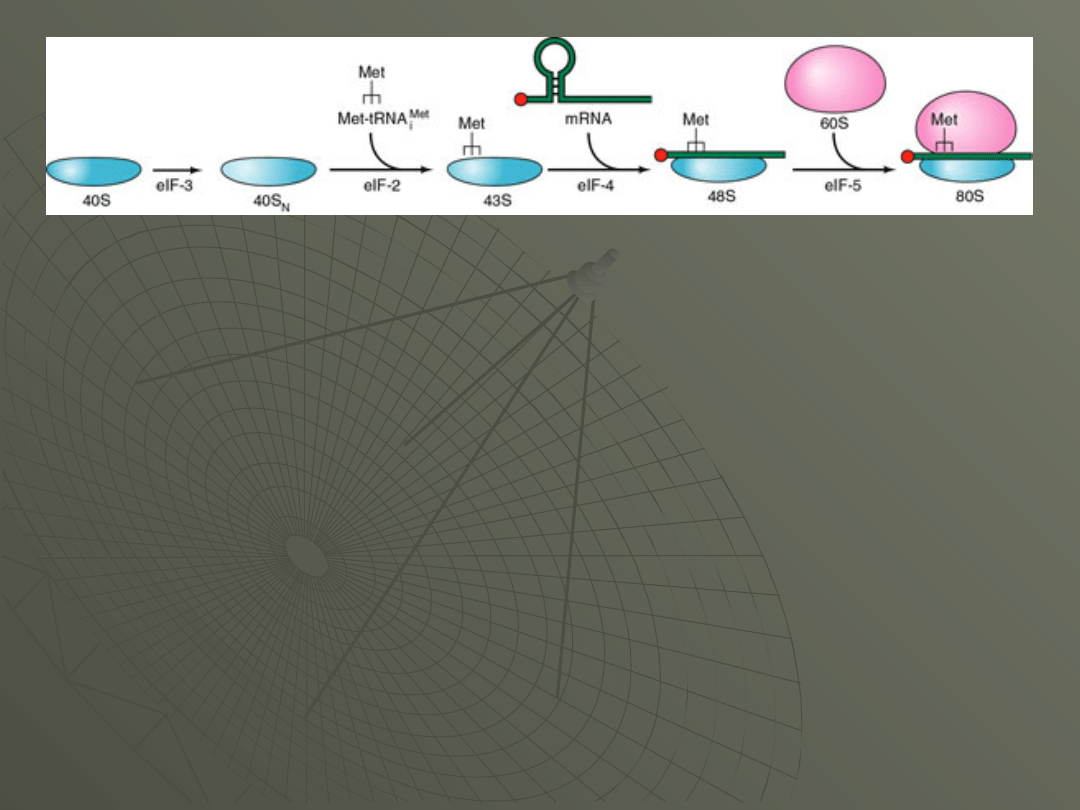
The 40S subunit joins with eIF-3 to form the
40SN structure that can join with the initiator
tRNA.
Along with eIF-2, the tRNA joins to form the
43S complex.
This now binds to the capped 5' end of mRNA,
under the direction if eIF-4, and scans down
the mRNA until it reaches the correct AUG
start codon.
This is the 48S initiation complex.
The 60S subunit now joins, using the action of
eIF-5, to form the final 80S initiation complex.

The unique event in this sequence that truly
distinguishes this from prokaryotic initiation is
the scanning of the 40S subunit.
This requires the presence of the 5'
methylated cap structure.
The initiation factor responsible for this
recognition is a factor called eIF-4F.
This consists of three separate proteins:
-eIF-4E (the cap binding protein itself),
-eIF-4A and
-eIF-4G.
All three together are eIF-4F.
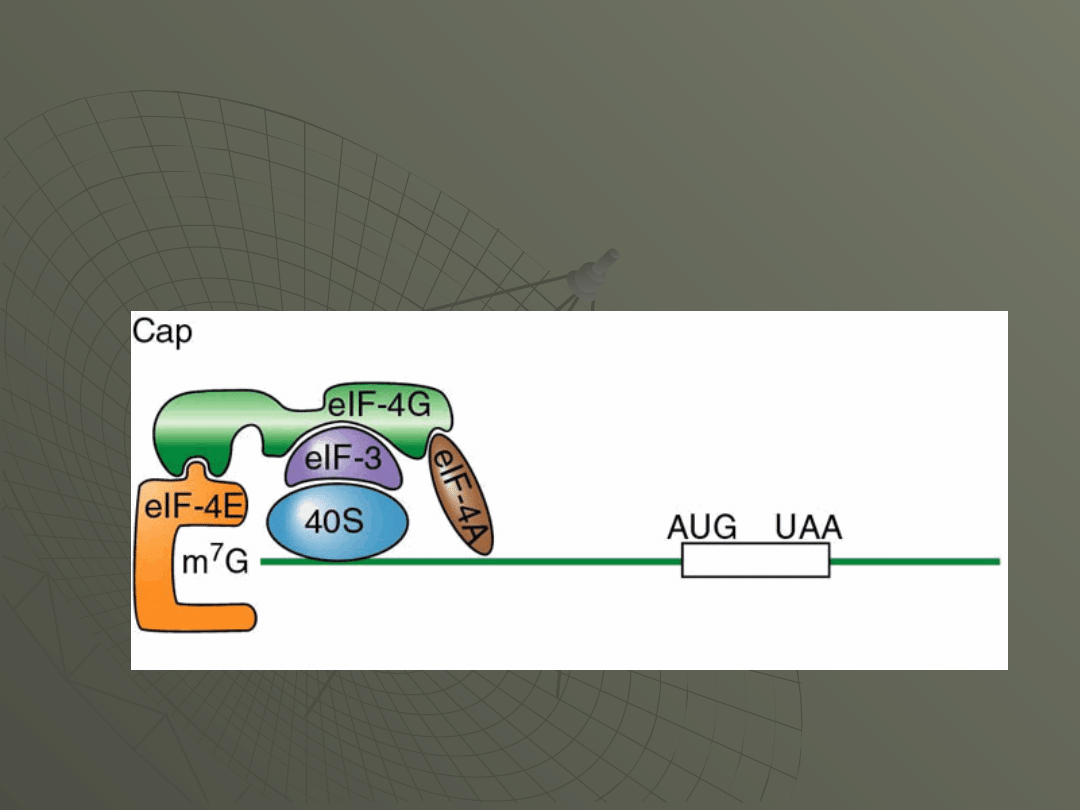
In Figure, this is shown as simply eIF-4, since
it is a complex of proteins.
Here's how it is thought these factors
interact at the cap:

The best evidence that the methylated cap is a
necessity for translation comes from infection of
cells with poliovirus.
This virus has an RNA genome that serves as a
messenger RNA inside the cell.
The genomic RNA of poliovirus is not capped
and methylated, and yet is translated in the
infected cell very efficiently.
It turns out that shortly after infection,
poliovirus shuts down the cell's ability to
translate any capped mRNA.
It does this by destroying eIF-4G, the stabilizing
protein of the cap binding complex eIF-4F.
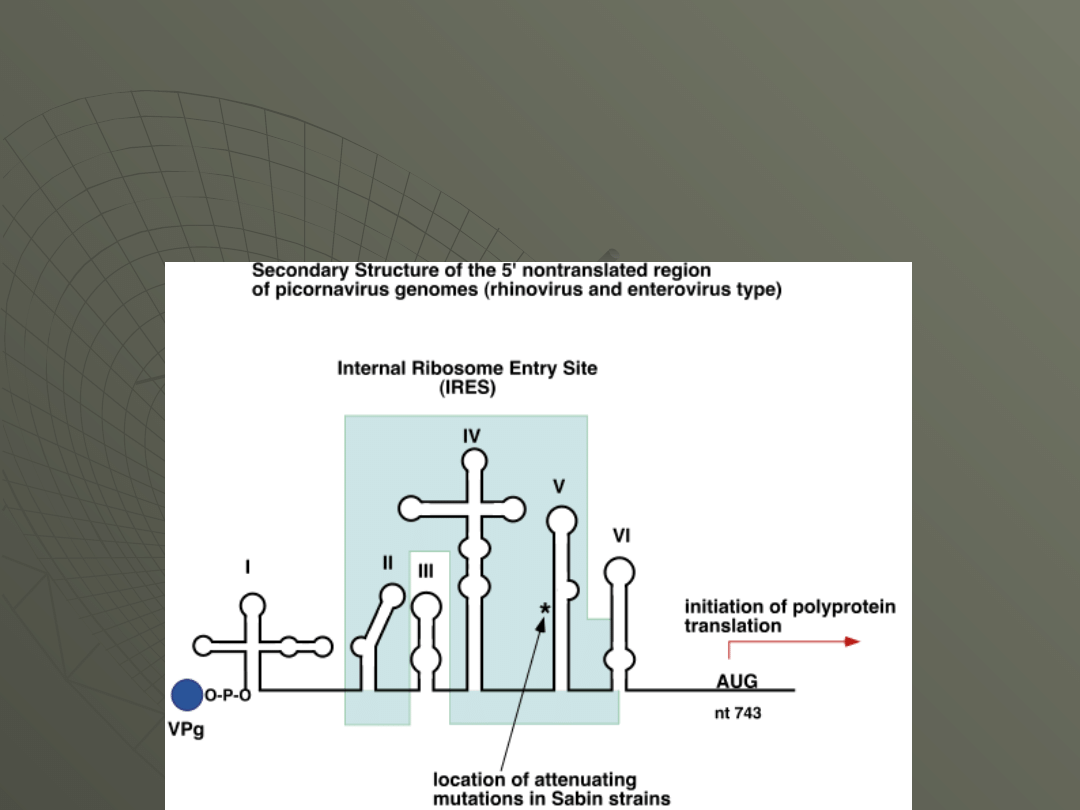
How is poliovirus RNA translated if it has no
cap? The viral RNA has an internal ribosome
entry site (IRES), a secondary structure feature
of the RNA that allows direct ribosome binding,
similar to that found in prokaryotes. Here's a
diagram of the poliovirus IRES:
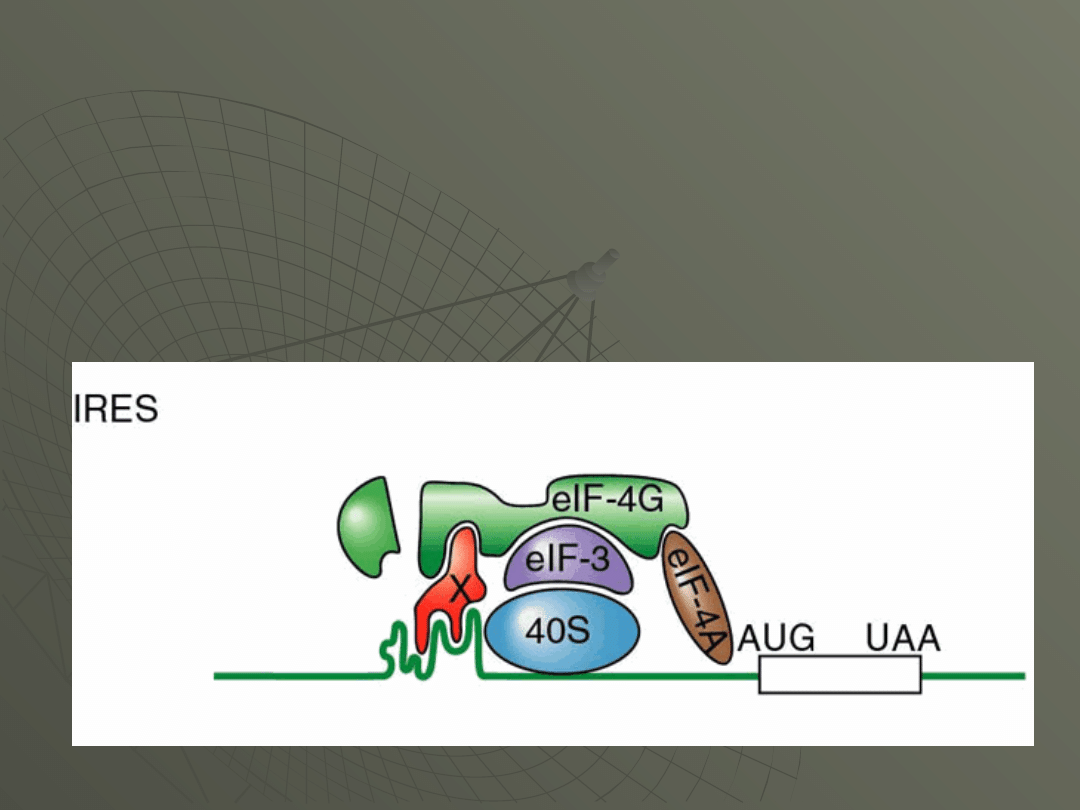
This feature becomes the place where the 40S
complex binds. The Figure is supposed to
show the situation in poliovirus-infected cell,
with a protein factor called "X" binding to the
IRES.
There is clear evidence in the literature that
documents the existence of this protein.
Document Outline
- Slide 1
- Slide 2
- Slide 3
- Slide 4
- Slide 5
- Slide 6
- Slide 7
- Slide 8
- Slide 9
- Slide 10
- Slide 11
- Slide 12
- Slide 13
- Slide 14
- Slide 15
- Slide 16
- Slide 17
- Slide 18
- Slide 19
- Slide 20
- Slide 21
- Slide 22
- Slide 23
- Slide 24
- Slide 25
- Slide 26
- Slide 27
- Slide 28
- Slide 29
- Slide 30
- Slide 31
- Slide 32
- Slide 33
- Slide 34
- Slide 35
- Slide 36
Wyszukiwarka
Podobne podstrony:
2014 BPEG część 5 inicjacja translacji
33 Przebieg i regulacja procesu translacji
BM7 Translacja
Inicjacja seksualna młodzieży gimnazjalnej na przykładzie szkoły wiejskiej
Ekspresja informacji genetycznej-transkrypcja i translacja, NAUKA
In literary studies literary translation is a term of two meanings rev ag
TRANSLATORIUM KOLOKWIUM !!!
inicjatywa ustawodawcza referat
Early Theories of Translation
Erratum to Revised Transliminality scale
historical identity of translation
Magiczna inicjacja
INICJACJA, Szamanizm, rozwoj duchowy
dokumenty-translation-all
Translatoryka bibliografia
Przeklad ogolny- zaliczenie, Filologia angielska, Notatki I rok, general translation
translantologia - odpowiedzi(1), SPECJALIZACJA pielęgniarstwo operacyjne, Testy
2012 i ogólnoplanetarna szyszynkowa inicjacja ludzkości, Medycyna, Szyszynka
więcej podobnych podstron