
C A N C E R
G E N E T I C S
ALEKSANDER L. SIEROŃ
KATEDRA I ZAKŁAD
BIOLOGII OGÓLNEJ MOLEKULARNEJ I GENETYKI
ŚLĄSKA AKADEMIA MEDYCZNA
*** KATOWICE 2006***
http://biolmolgen.slam.katowice.pl
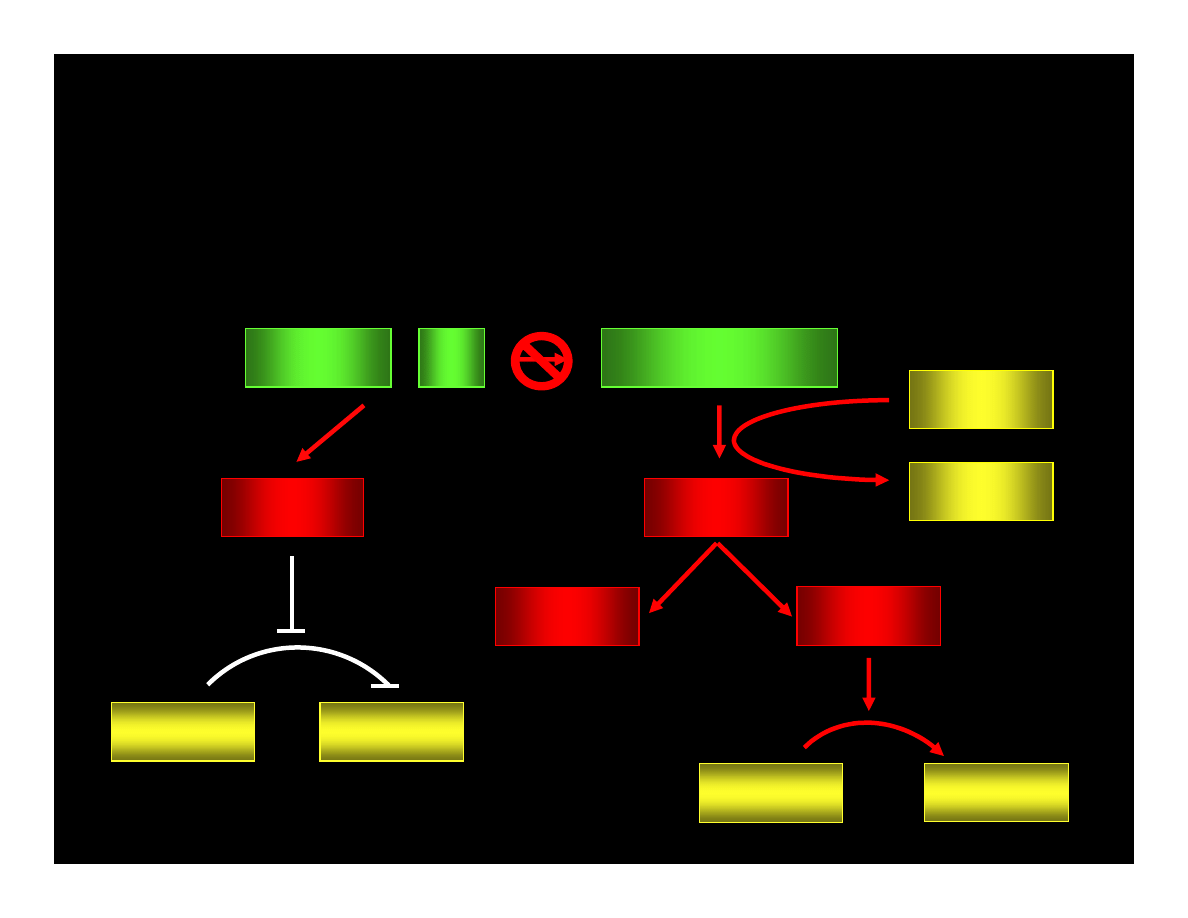
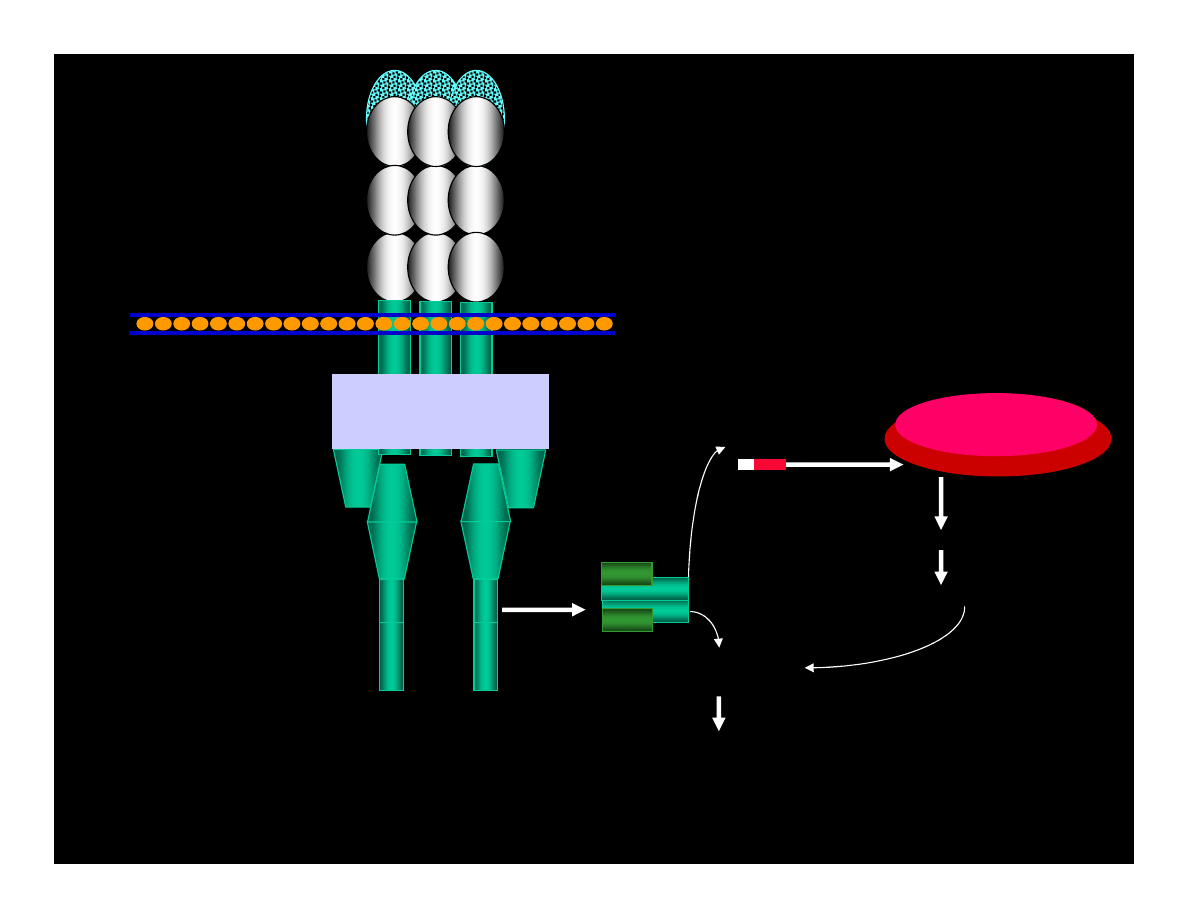
CYCLIN : Cdk
pRB : E2F
ppRB : E2F
E2F
ATP
pRB : E2F
ADP
PHASE G1
PHASE S
CYCLIN
Cdk
PHASE G1
PHASE S
REPAIRE OR
APOPTOSIS
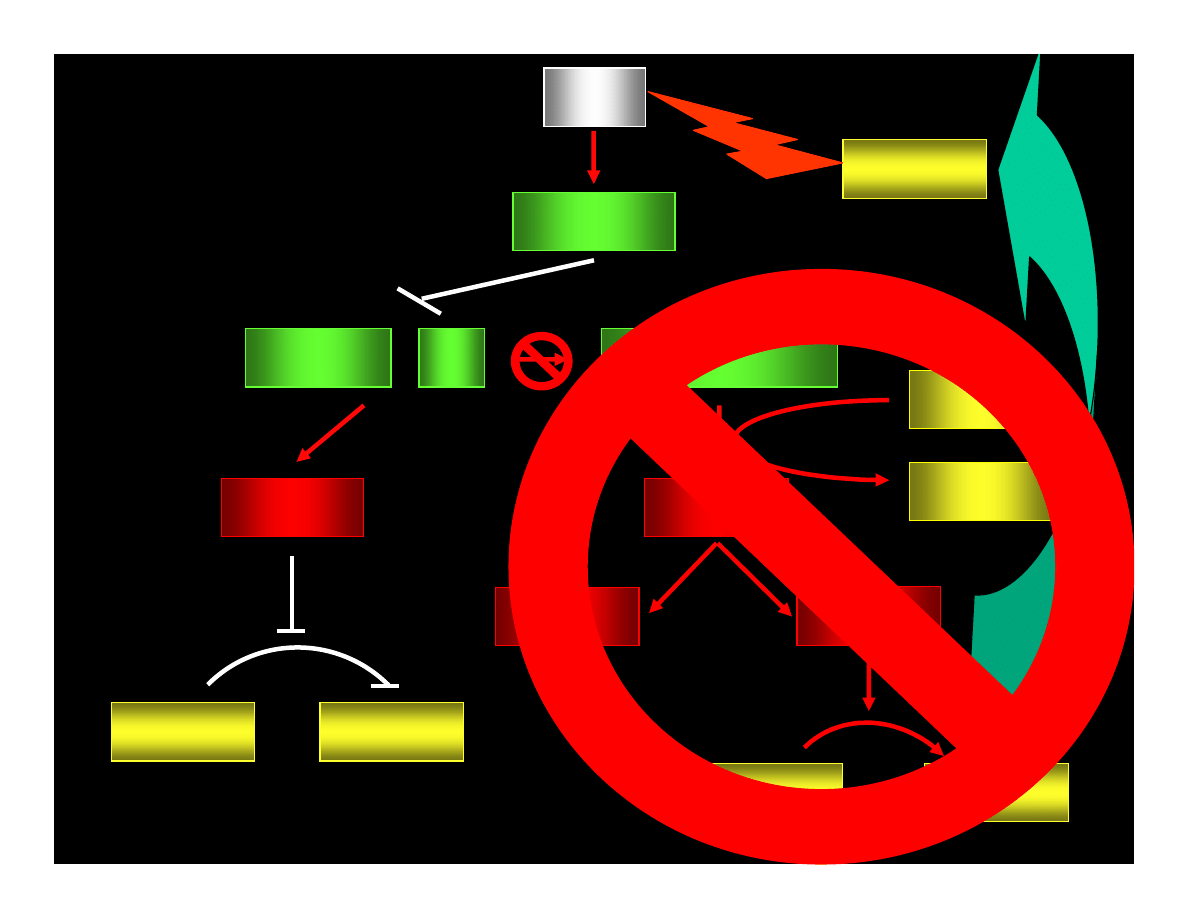
p53
p21
WAF1/CIP
CYCLIN : Cdk
pRB : E2F
ppRB : E2F
E2F
ATP
pRB : E2F
ADP
PHASE G1
PHASE S
CYCLIN
Cdk
PHASE G1
PHASE S
REPAIRE OR
APOPTOSIS
ERROR
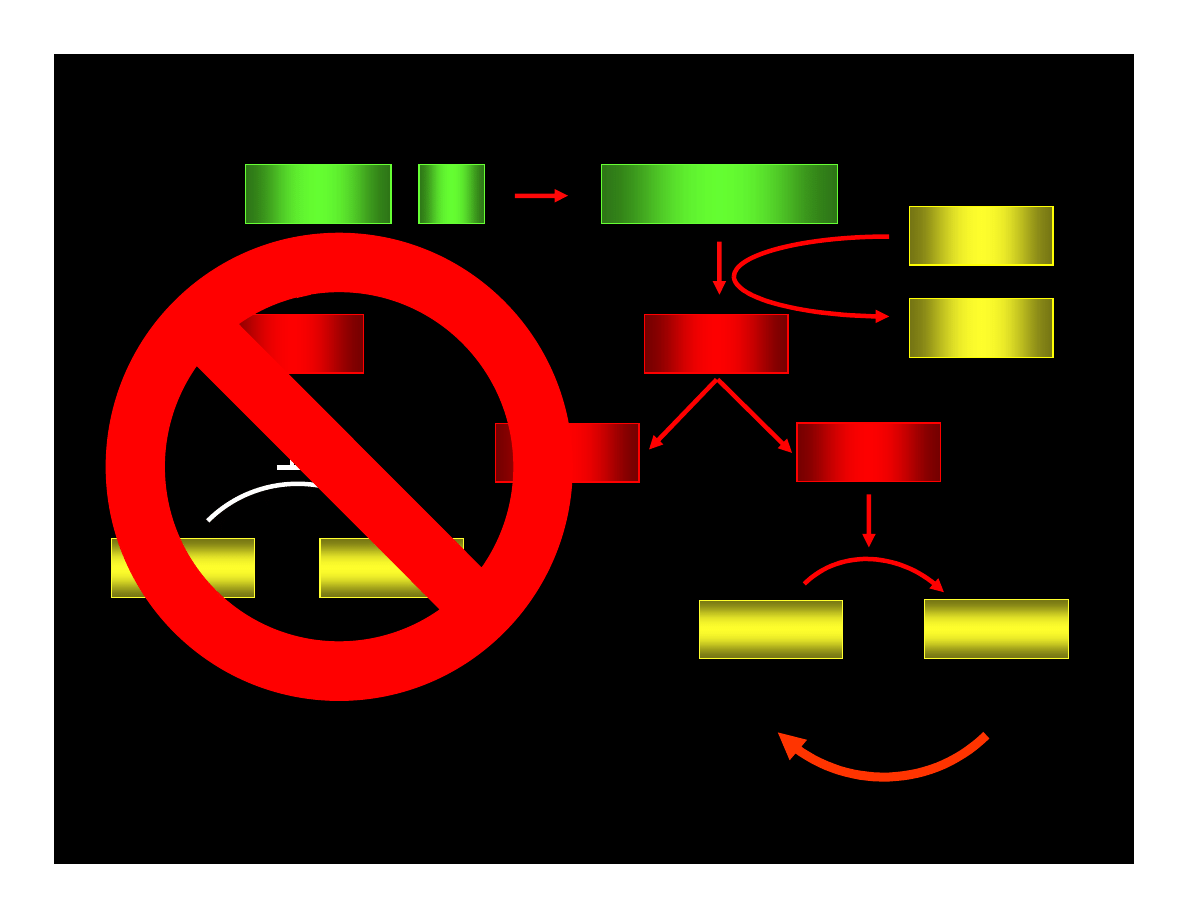
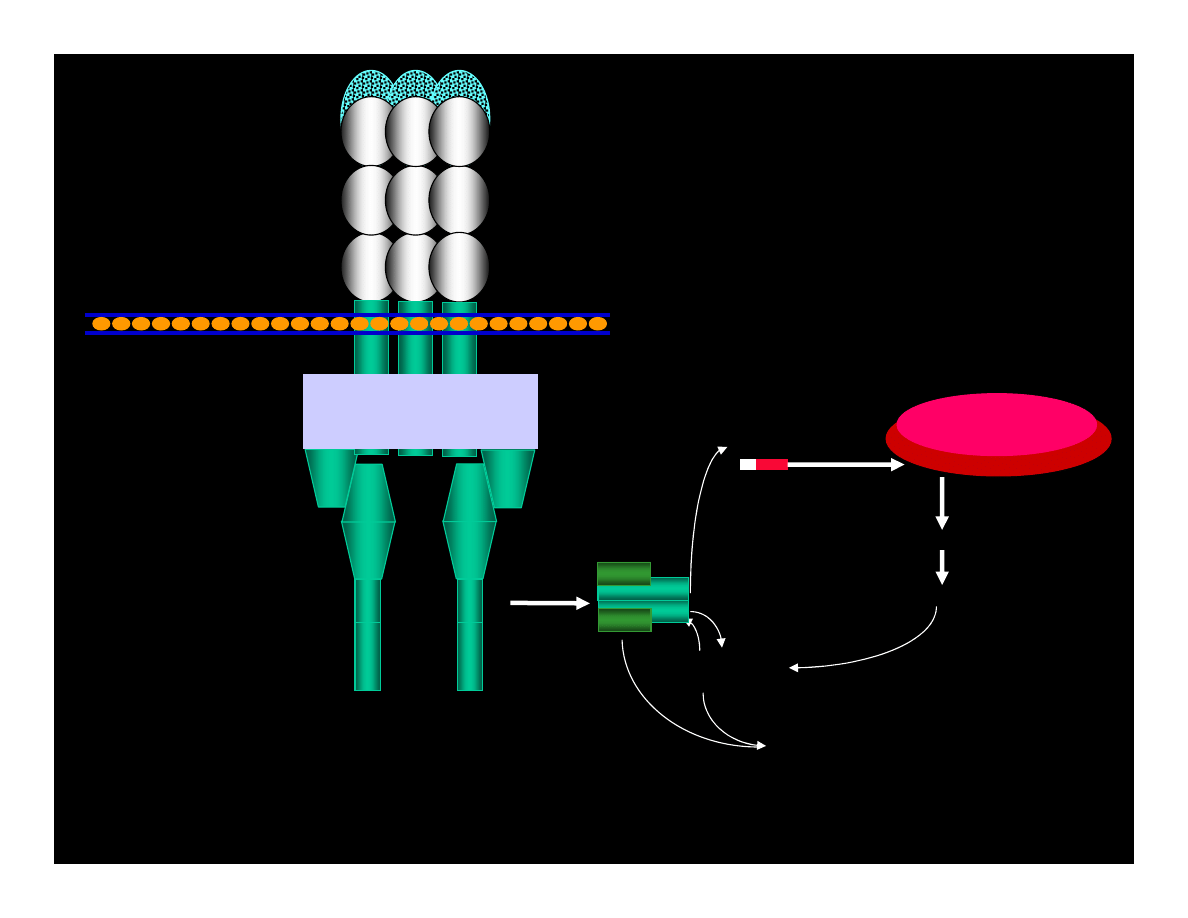
CYCLIN : Cdk
pRB : E2F
ppRB : E2F
E2F
ATP
pRB : E2F
ADP
PHASE G1
PHASE S
CYCLIN
Cdk
PHASE G1
PHASE S
REPAIRE OR
APOPTOSIS
IF LACK OF p53 ACTIVITY
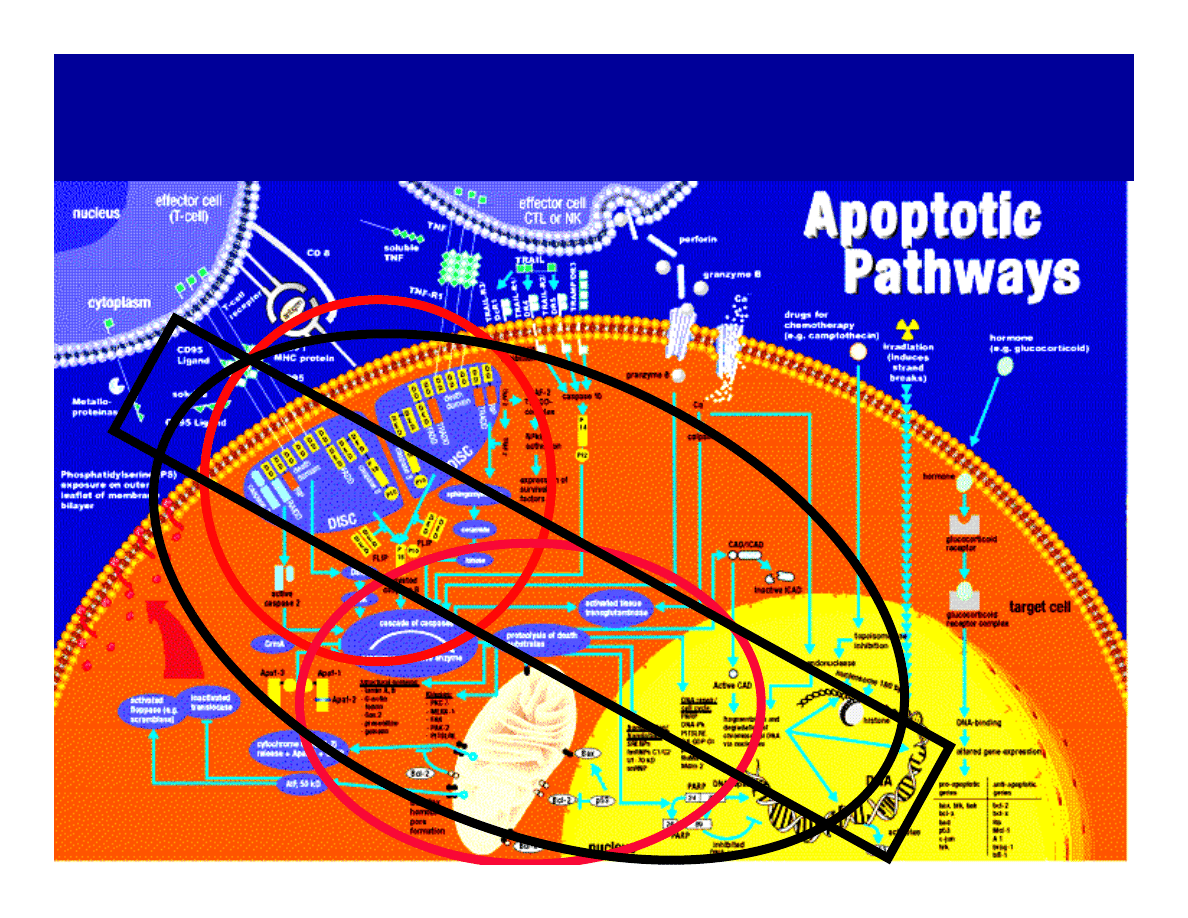
RECEPTOR
MITOCHONDRIAL
A P
O P
T O
S I
S
T
R
A
N
S
M
E
M
B
R
A
N
E
D
O
M
A
IN
EX
TR
AC
EL
LU
LA
R D
OM
AIN
C
Y
T
O
P
L
A
S
M
IC
D
O
M
A
IN
Complex
Complex
Complex
Complex
E
X
T
R
A
C
E
L
L
U
L
A
R
S
P
A
C
E
C
Y
T
O
P
L
A
S
M
FasL
Extracellular
Domain
pro-Caspase-8
pro-Caspase-8
pro--Bid
cytochrome c
Caspase-9/cytochrome c
Executing Caspases &
Other Substrates
Mitochondria
p15tBid
Fas/CD95
FADD
FADD
Cell Membrane
Receptor Pathway
Mitochondrial Pathway
(Cys-Proteinases)
INFLAM
ATION
A P
O P
T O
S I
S
pro--Bid
pro-Caspase-8
pro-Caspase-8
Cytochrome c
Caspase-9/cytochrome c
Executing Caspases &
Other Substrates
Mitochondria
p15tBid
Extracellular
Domain
FasL
Fas/CD95
Cell Membrane
FADD
FADD
Caspase 8
Receptor Pathway
Mitochondrial Pathway
After Ligand Binding (e.g.: Fas)
Execyting Caspases
& Other Substrates
Caspase 3
pro-Caspase-8
pro-Caspase-8
pro--Bid
Caspase 8
Cytochrome c
Caspase-9/cytochrome c
Mitochondria
p15tBid
Extracellular Domain
FasL
Fas/CD95
Cell Membrane
FADD
FADD
Receptor Pathway
Mitochondrial Pathway
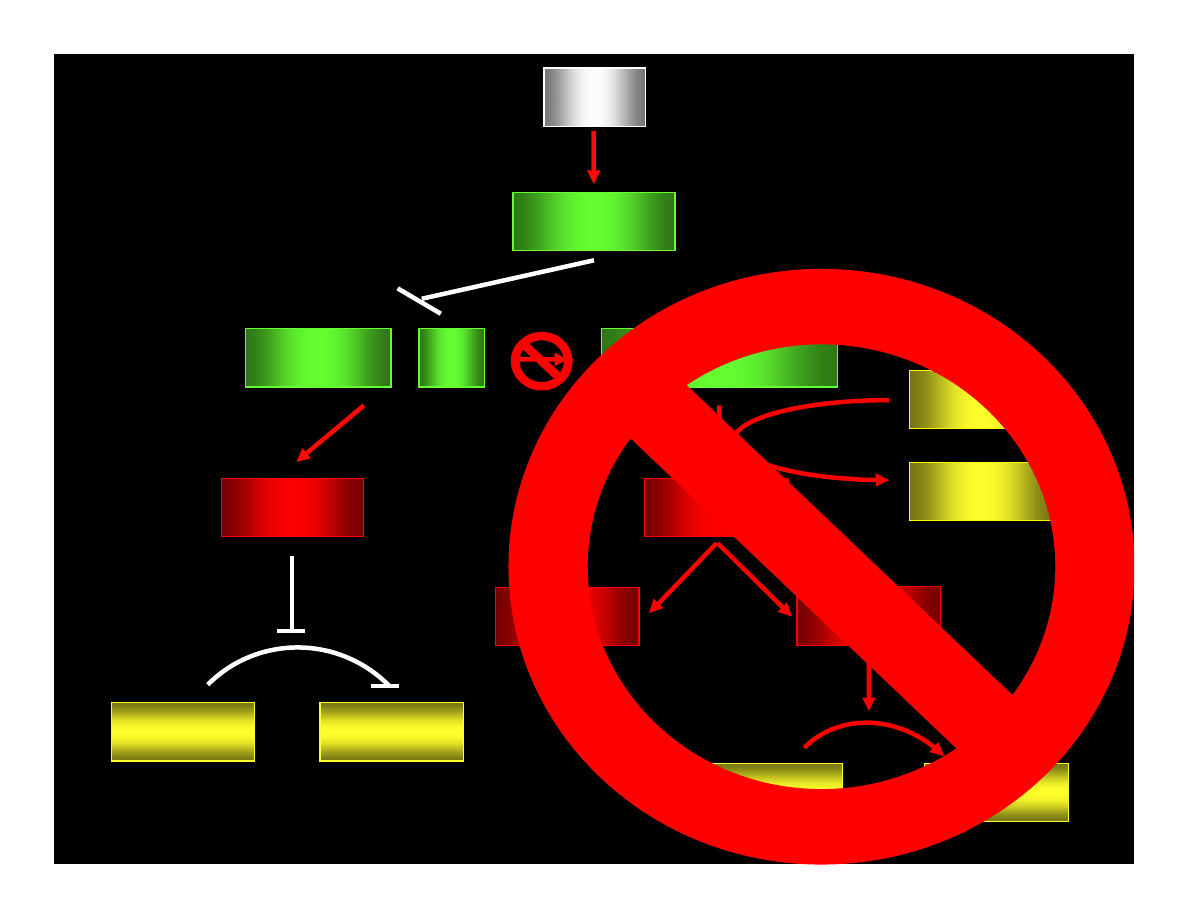
MUTATIONS
of
A P O P T O S IS
REGULATORS
DiSTU
rbaNc
e
T
ro
uG
h
&
CELL CYCLE
REGULATORS
pro-Caspase-8
pro-Caspase-8
Extracellular
Domain
FasL
Fas/CD95
Cell Membrane
FADD
FADD
MUTATIONS
INACTIVE
CASPASE 8
Executing Caspases &
Other Substrates
Caspase 3
Cytochrome c
Caspase-9/cytochrome c
Mitochondria
p15tBid
Nucleus
B
cl
2
MUTATIONS
INACTIVE
RECEPTORS
MUTATIONS INACTIVE LIGANDS
MUTATIONS
INACTIVE
KINASES
=
???N
EOPL
AST
IC T
RAN
SFO
RMA
TIO
N???
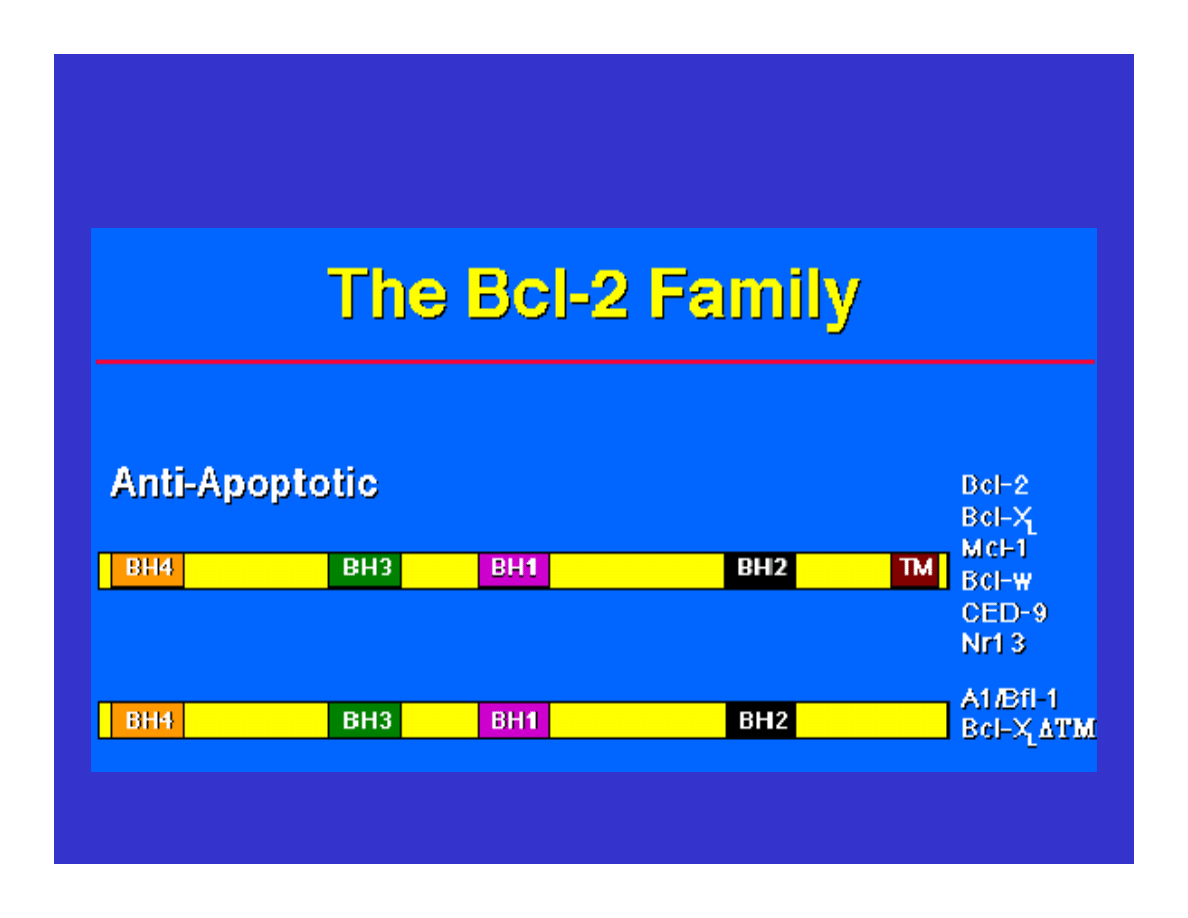
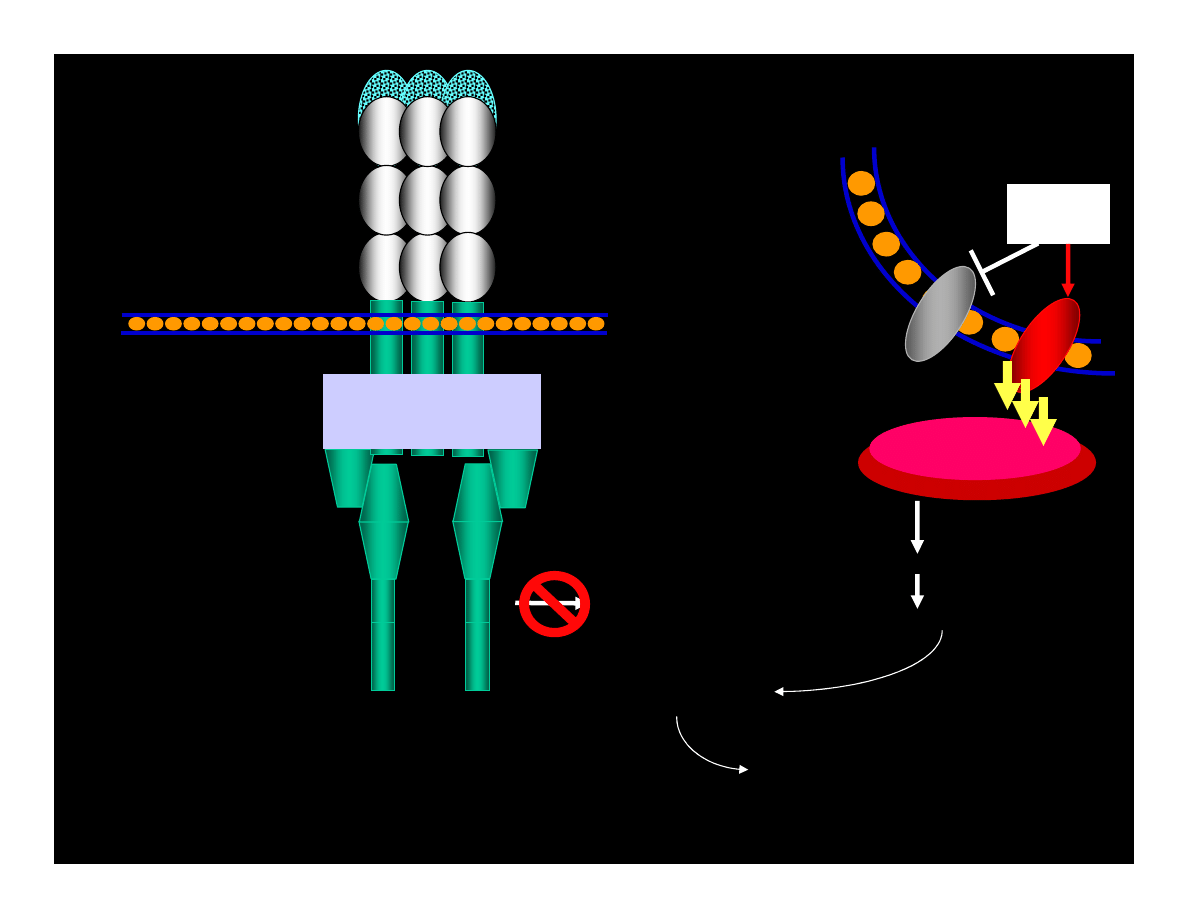
Blocking Apoptosis
(Eight known proteins in 2001)
pro-Caspase-8
pro-Caspase-8
Extracellular
Domain
FasL
Fas/CD95
Błona komórkowa
FADD
FADD
Nucleus
B
cl
2
MUTATIONS
INACTIVE
CASPASE 8
Executing Caspases &
Other Substrates
Caspase 3
Cytochrome c
Caspase-9/Cytochrome c
p15tBid
Mitochondria
p53
B
ax
MUTATIONS
INACTIVE
RECEPTORS
MUTATIONS INACTIVE LIGANDS
MUTATIONS
INACTIVE
KINASES
p53
p21
WAF1/CIP
CYCLIN : Cdk
pRB : E2F
ppRB : E2F
E2F
ATP
pRB : E2F
ADP
PHASE G1
PHASE S
CYCLIN
Cdk
PHASE G1
PHASE S
REPAIR OR
APOPTOSIS
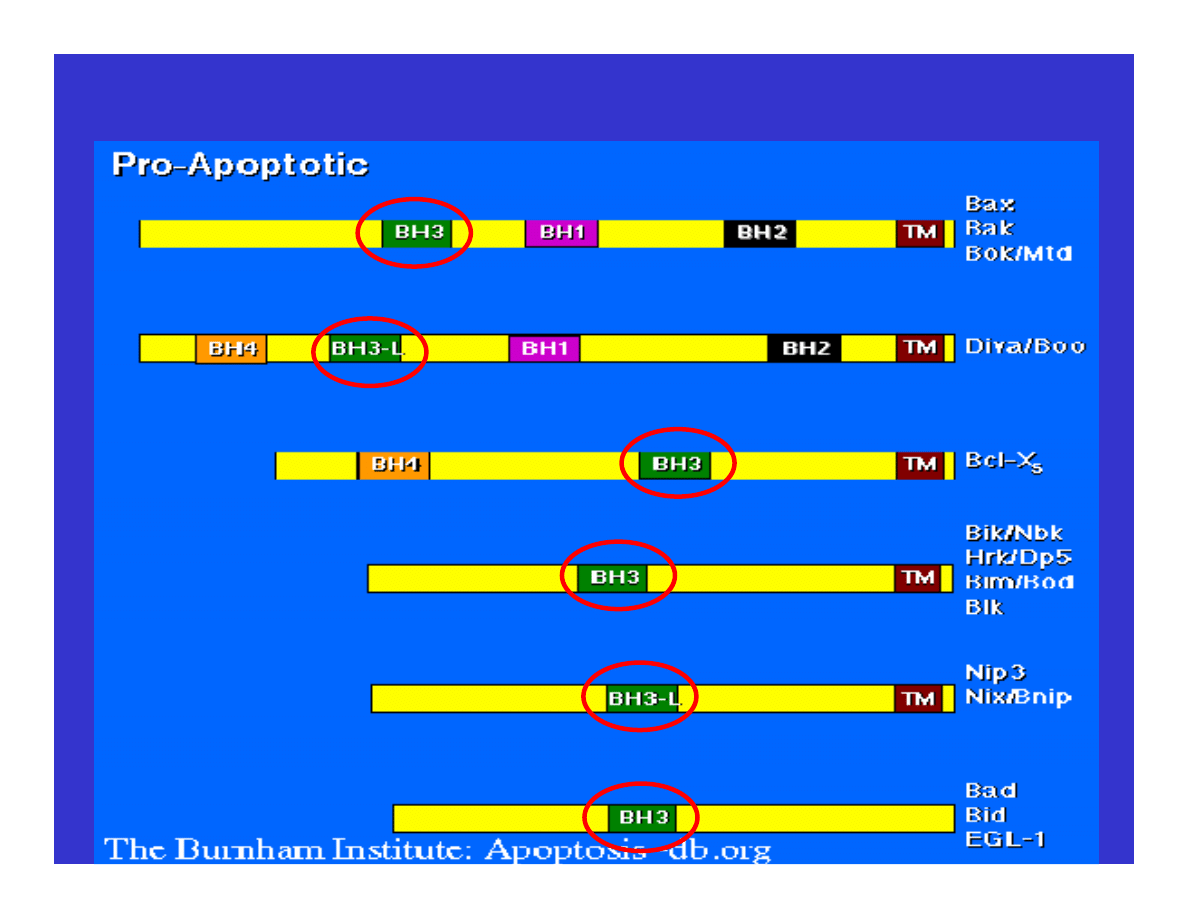
Induction of Apoptosis
(fourteen proteins known in 2001)
pro-Caspase-8
pro-Caspase-8
Extracellular
Domain
FasL
Fas/CD95
Cell Membrane
FADD
FADD
Nucleus
B
cl
2
MUTATIONS
INACTIVE
CASPASE 8
Mitochondria
p53
B
ax
MU
TA
TI
ON
S
M
U
T
A
T
IO
N
S
!!!N
EOP
LAS
TIC
TR
AN
SFO
RM
ATI
ON
!!!
MU
TA
TIO
NS
Complex
Complex
E
X
T
R
A
C
E
L
L
U
L
A
R
C
Y
T
O
P
L
A
S
M
Complex
Complex
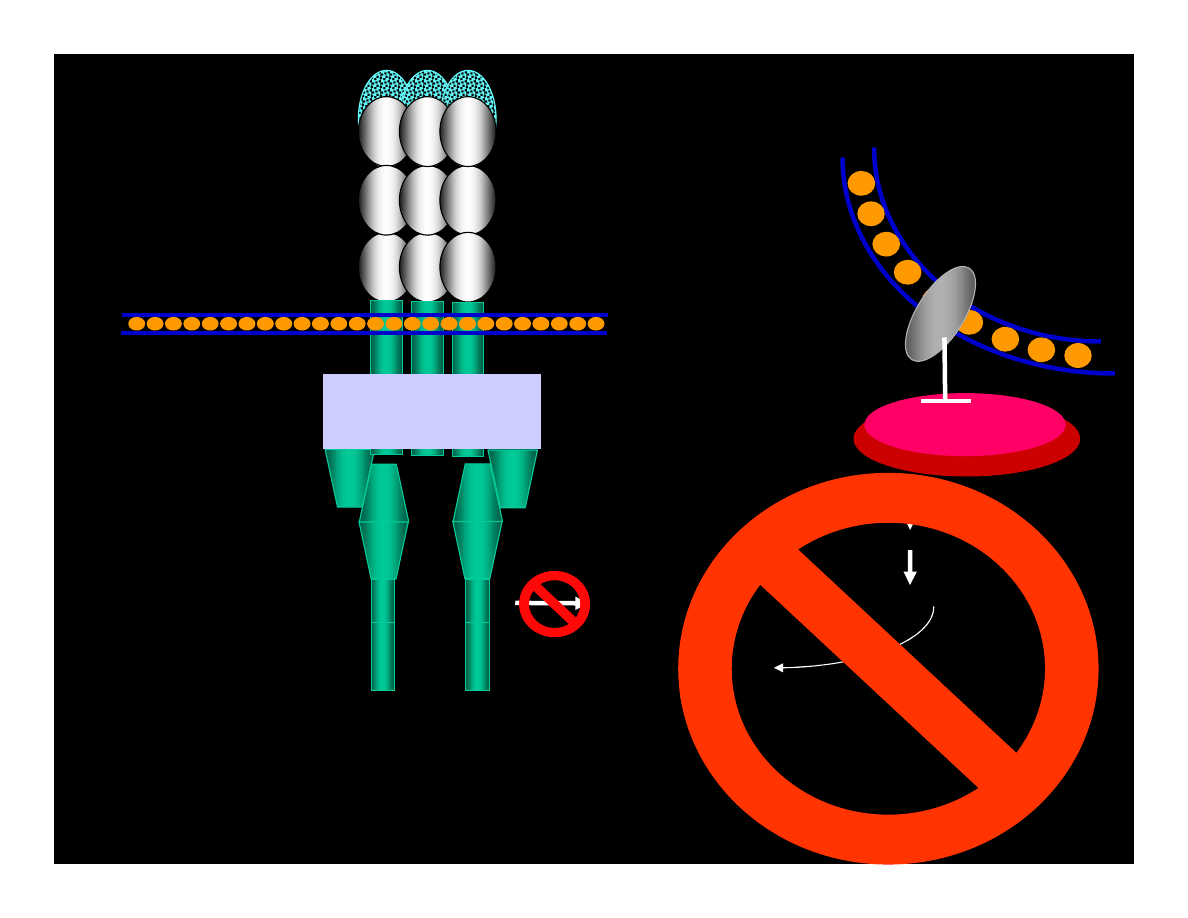
(Wyniki mutacji)
NO
REKRUTATION
OF
DEATH
COMPLEX
NO
PROTEOLYTIC
ACTIVITY
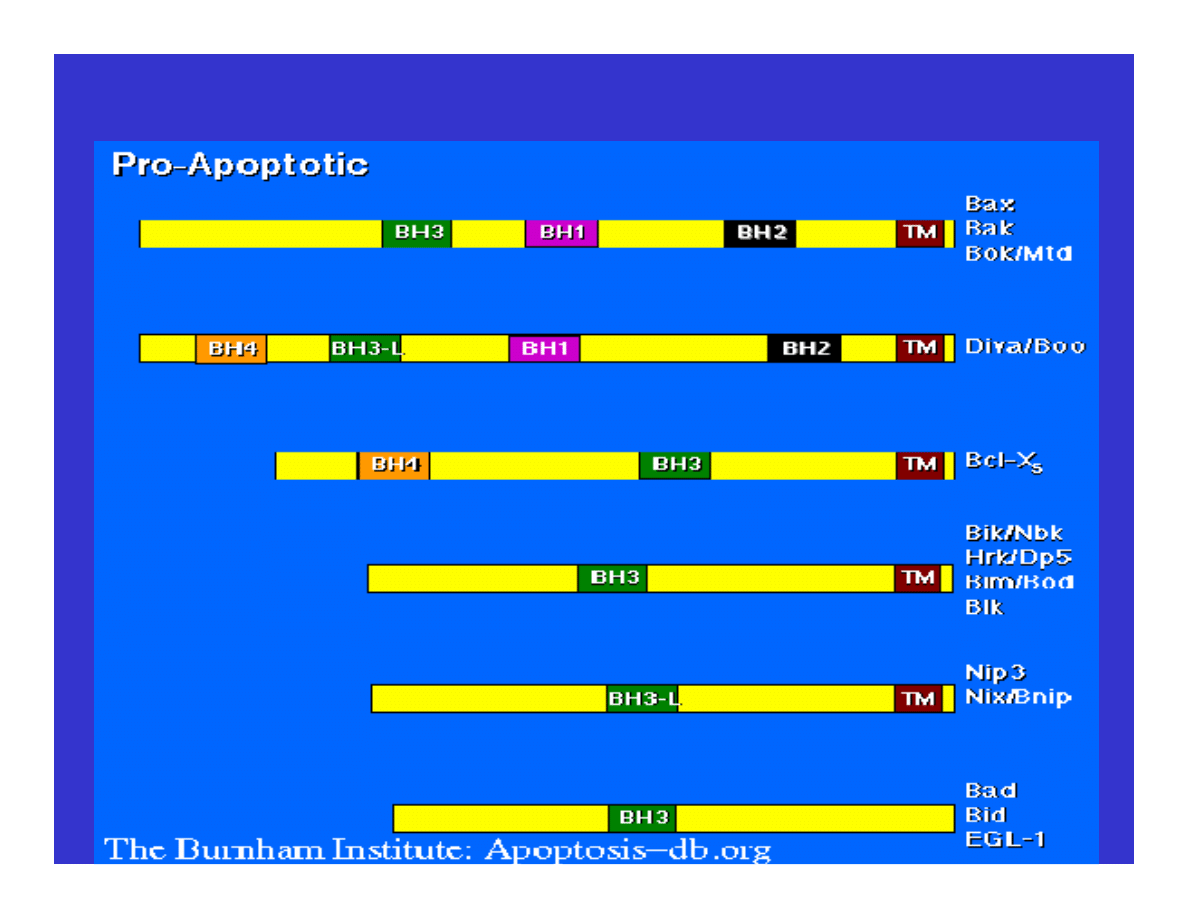
Mutations of Apoptosis Inducing Factors
***
***
***
***
***
***
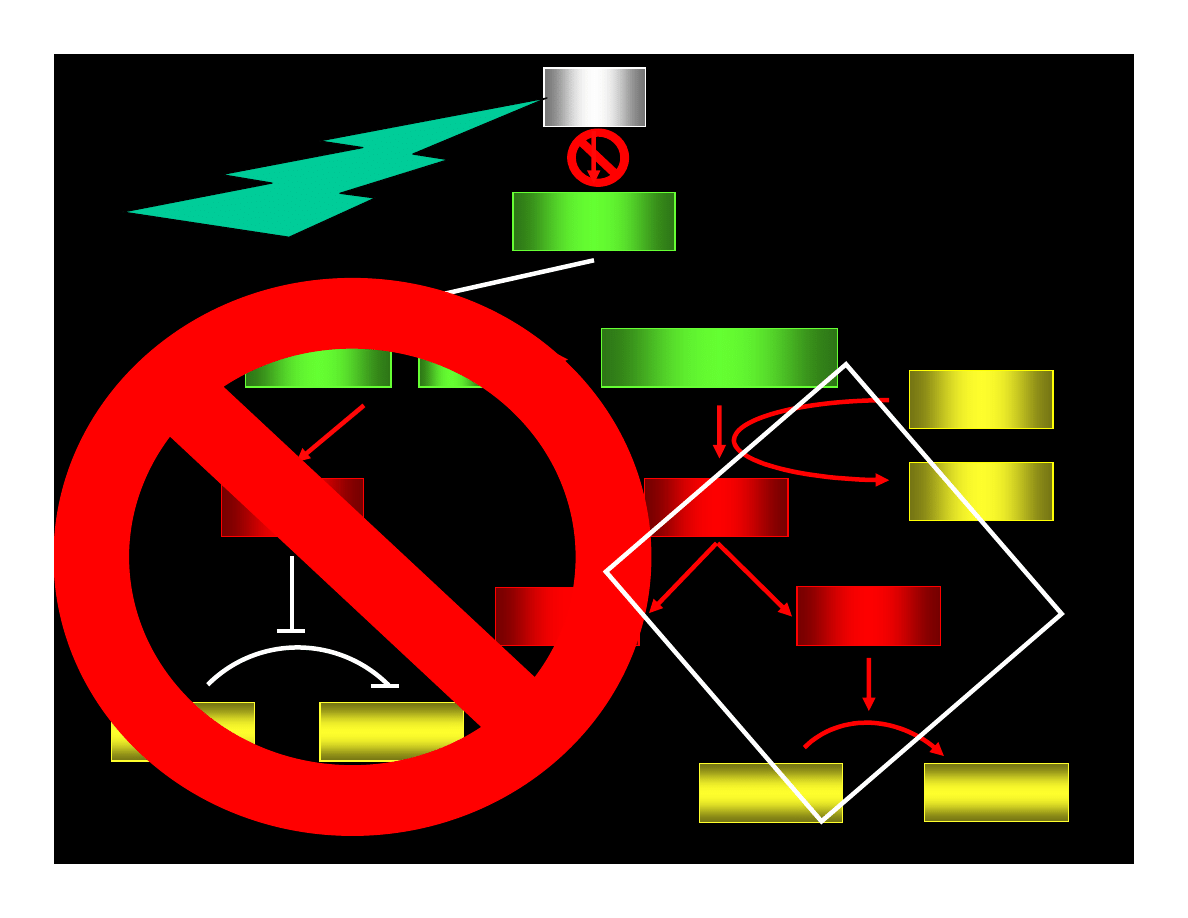
p53
p21
WAF1/CIP
CYCLIN : Cdk
pRB : E2F
ppRB : E2F
E2F
ATP
pRB : E2F
ADP
PHASE G1
PHASE S
CYCLIN
Cdk
PHASE G1
PHASE S
REPAIR OR
APOPTOSIS
MUTATIONS
U
N
CO
N
TR
O
LE
D
D
IV
IS
II
O
N
S
CU
M
U
LA
TI
O
N
O
F
M
U
TA
TI
O
N
S
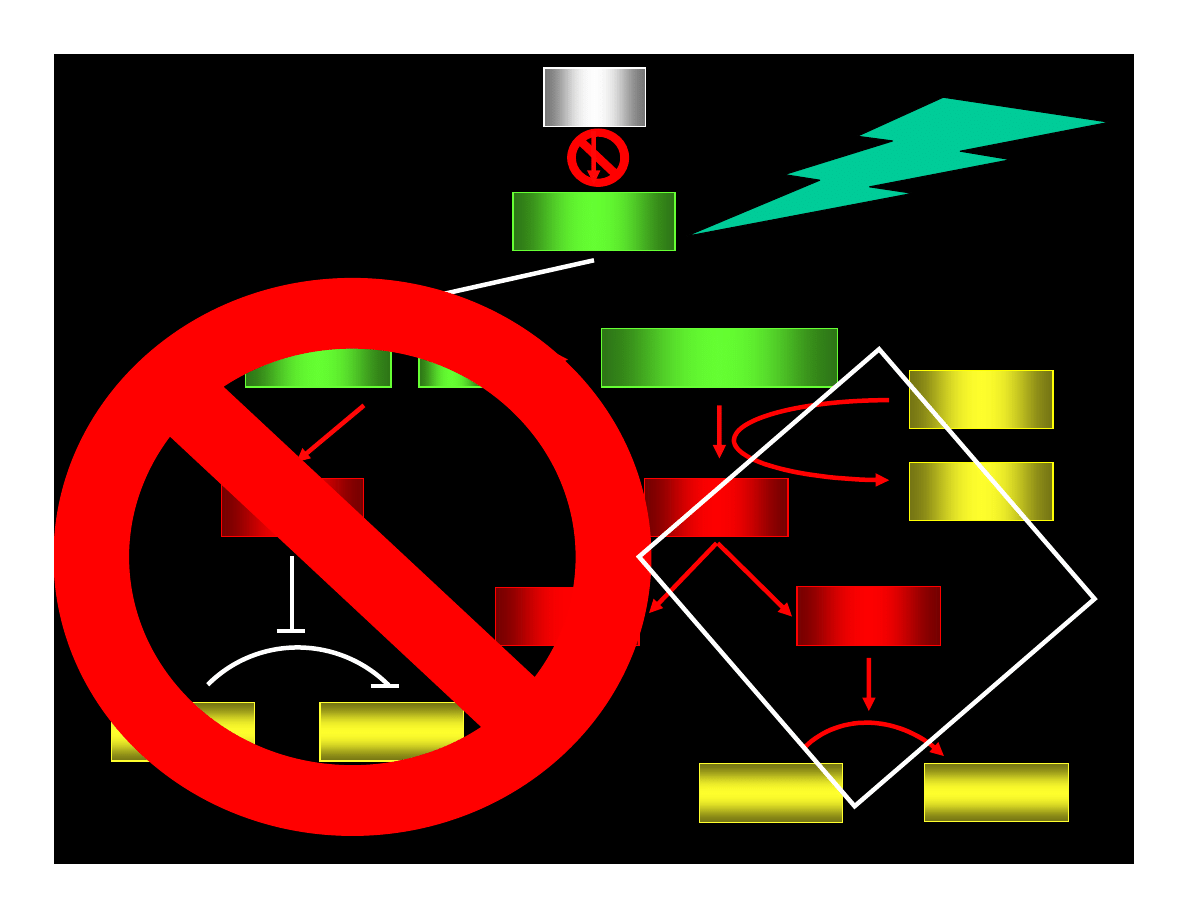
p53
p21
WAF1/CIP
CYCLIN : Cdk
pRB : E2F
ppRB : E2F
E2F
ATP
pRB : E2F
ADP
PHASE G1
PHASE S
CYCLIN
Cdk
PHASE G1
PHASE S
REPAIR OR
APOPTOSIS
MUTATIONS
U
N
CO
N
TR
O
LE
D
D
IV
IS
IO
N
S
CU
M
U
LA
TI
O
N
O
F
M
U
TA
TI
O
N
S
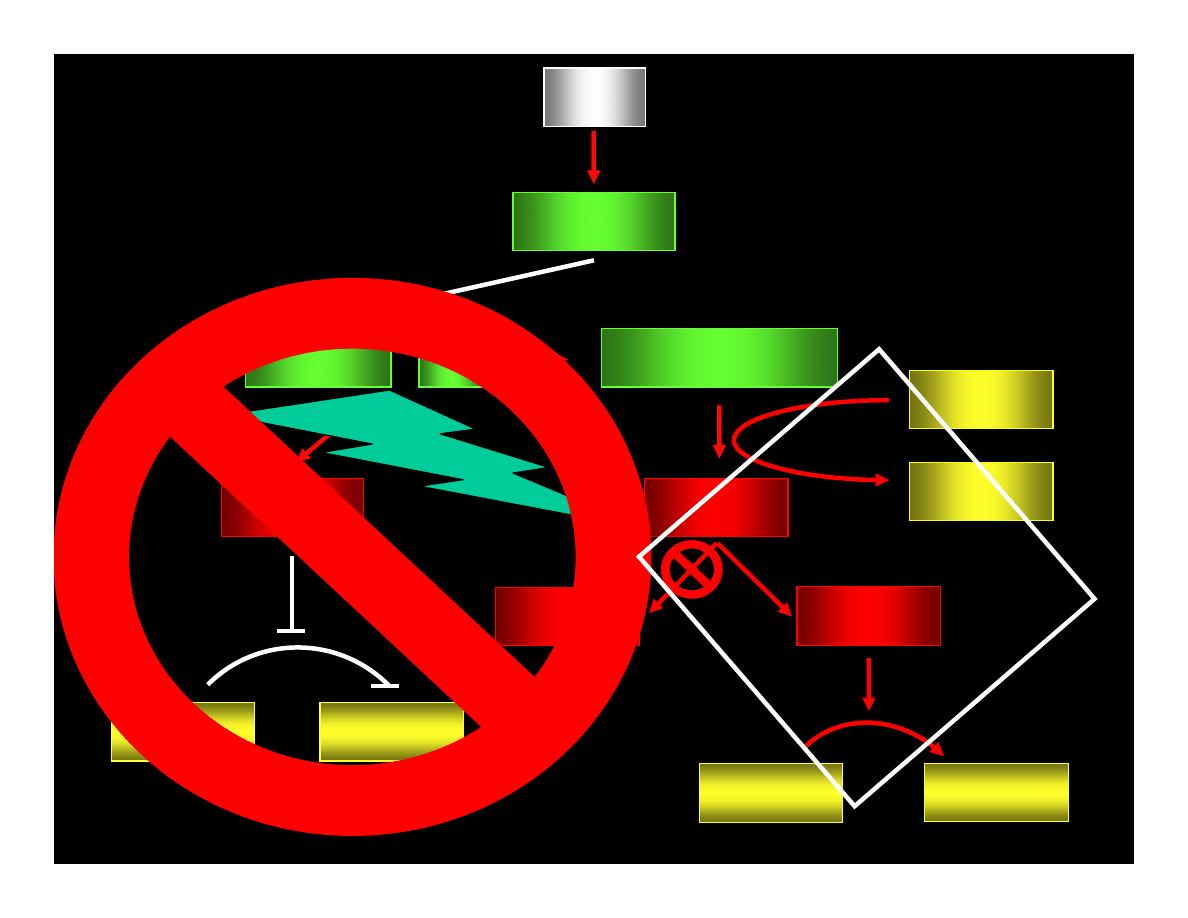
p53
p21
WAF1/CIP
CYCLIN : Cdk
pRB : E2F
ppRB : E2F
E2F
ATP
pRB : E2F
ADP
PHASE G1
PHASE S
CYCLIN
Cdk
PHASE G1
PHASE S
REPAIR OR
APOPTOSIS
MUTATIONS
U
N
CO
N
TR
O
LE
D
D
IV
IS
IO
N
S
CU
M
U
LA
TI
O
N
O
F
M
U
TA
TI
O
N
S

FEW
EXAMPLES
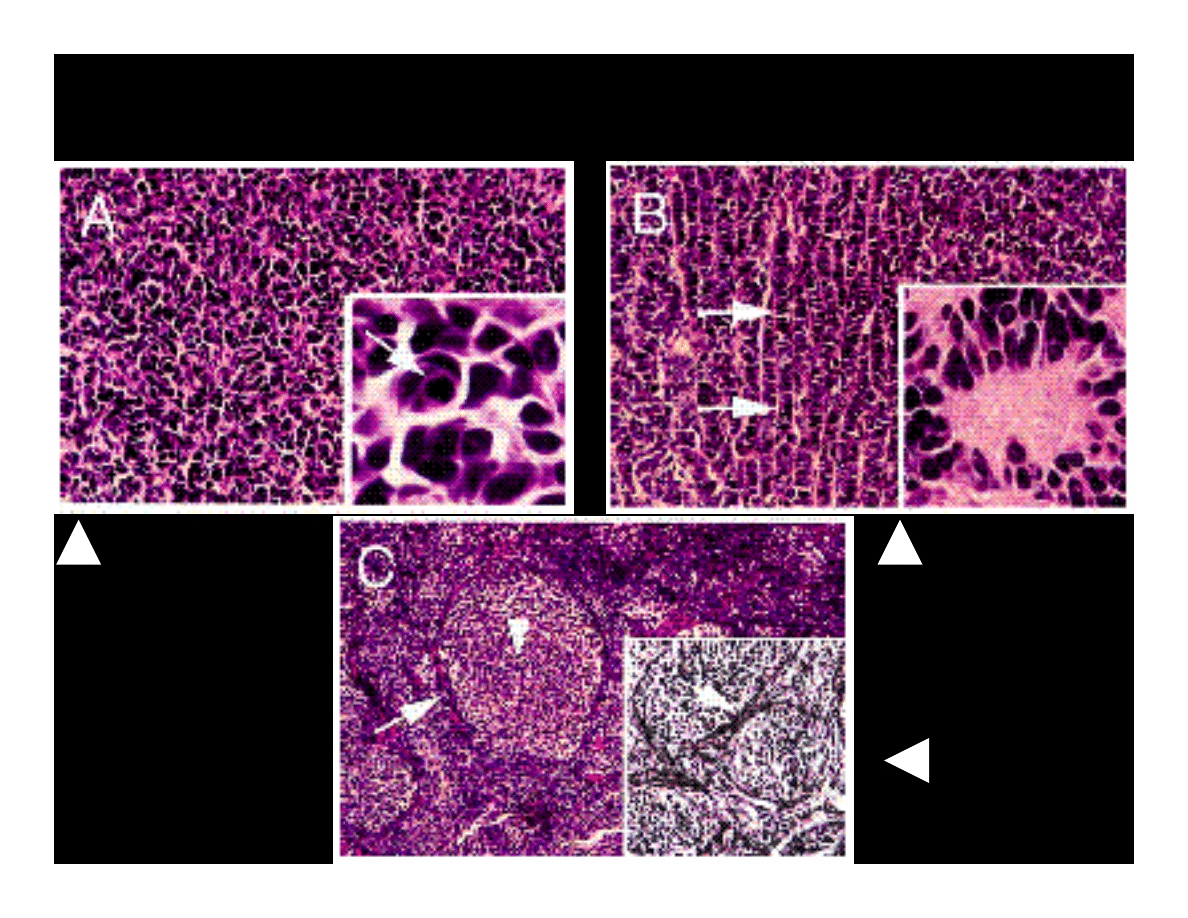
Clasic
Medulloblastoma
Neuroblastic
Medulloblastoma
Desmoplastic
Medulloblastoma
CLASSIFICATION OF
MEDULLOBLASTOMAS
Krynska B., et al.
(1999) PNAS,
96:11519-24
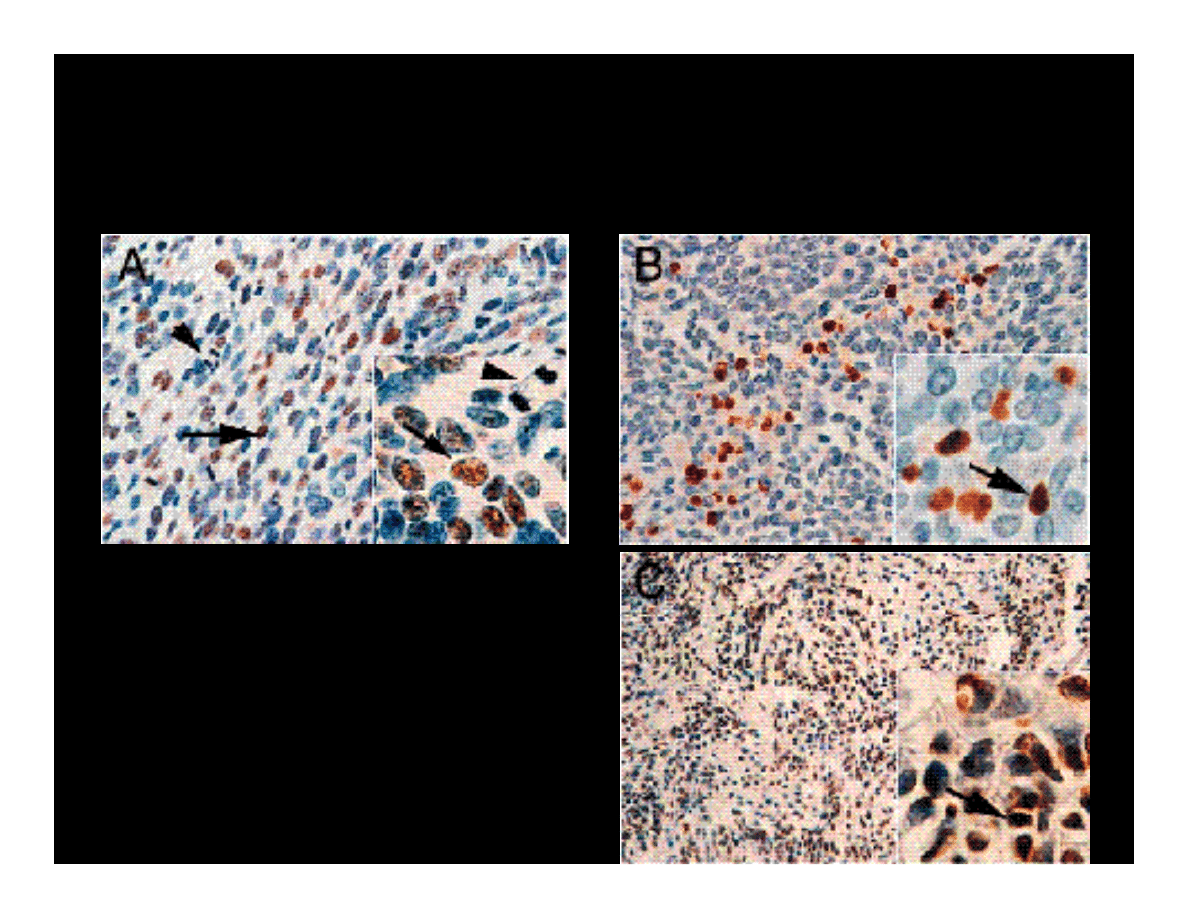
IMMUNOHISTOCHEMICAL DETECTION
OF JCV T-ANTIGEN IN MEDULLOBLASTOMAS
Magnification x400 (A &
B); x200 (C); x1000
(inserts)
Arrowheads point to
proliferating cells
Arrows indicate cells with
JCV T-antigen
K
ry
ns
ka
B.
,
et
a
l.
(1
9
9
9
)
PN
A
S
,
9
6
:1
15
19
-2
4
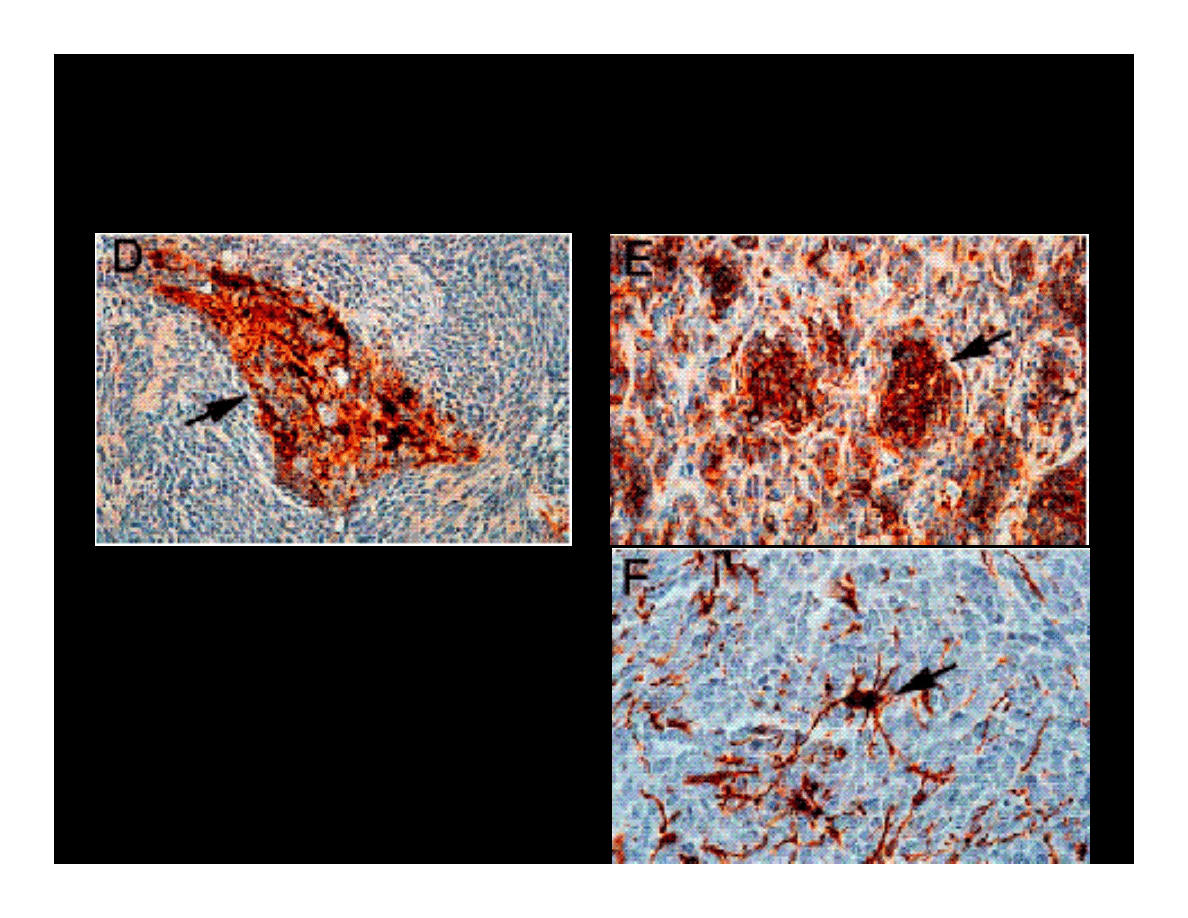
IMMUNOHISTOCHEMICAL ANALYSIS OF
MEDULLOBLASTOMA MARKERS
GFAP
Magnifications x200 (D) &
x400 (E i F).
Synaptophysin
ββββ
tubulin class III
K
ry
ns
ka
B.
,
et
a
l.
(1
9
9
9
)
PN
A
S
,
9
6
:1
15
19
-2
4
1469
2533
VP
1
47
71
44
26
5
0
1
3
2603
T
1
7
3
b
p
(4
3
0
3
-4
3
2
7
)
(4
2
5
5
-4
2
7
4
)
(4
40
8-
44
27
)
(2
03
9-
20
19
)
(1
8
4
8
-1
8
2
8
)
(1
89
1-
18
72
)
21
2
b
p
(27
97-2
776
)
(2600-2578)
(2668-26
63)
220 bp
PCR primers
Amplified DNA
Probe
JCV
0.6
0.7
0.8
0.9
0.0
0.1
0.2
0.3
0.4
0.5
(Mad-1)
5130 bp
Control Region
5130/0
44
95
2
7
7
4
9
2
5
2
6
88
3
1560
Ag
no
V
P
3
V
P
2
t
Krynska B., et al.
(1999) PNAS,
96:11519-24
JC VIRUS
GENOME
PCR DETECTION OF JCV DNA
IN MEDULLOBLASTOMAS
Early coding region
N-terminus of T-antigen
Early Coding Region,
C-terminus of T-antigen
Late coding region of VP1
capsid protein
Krynska B., et al.
(1999) PNAS,
96:11519-24
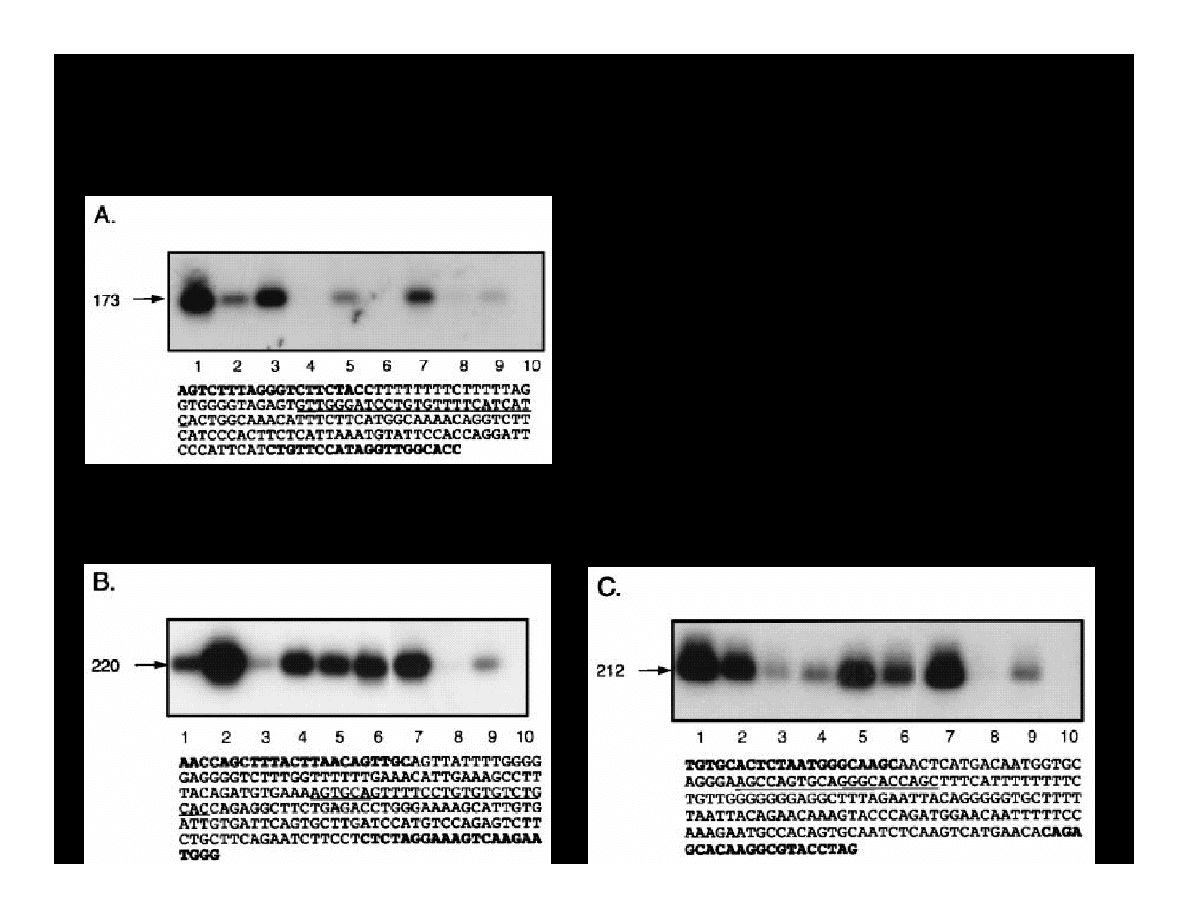
Krynska B., et al. (1997) JCB, 67:223-30
T ANTIGEN & p53 COMPLEXES
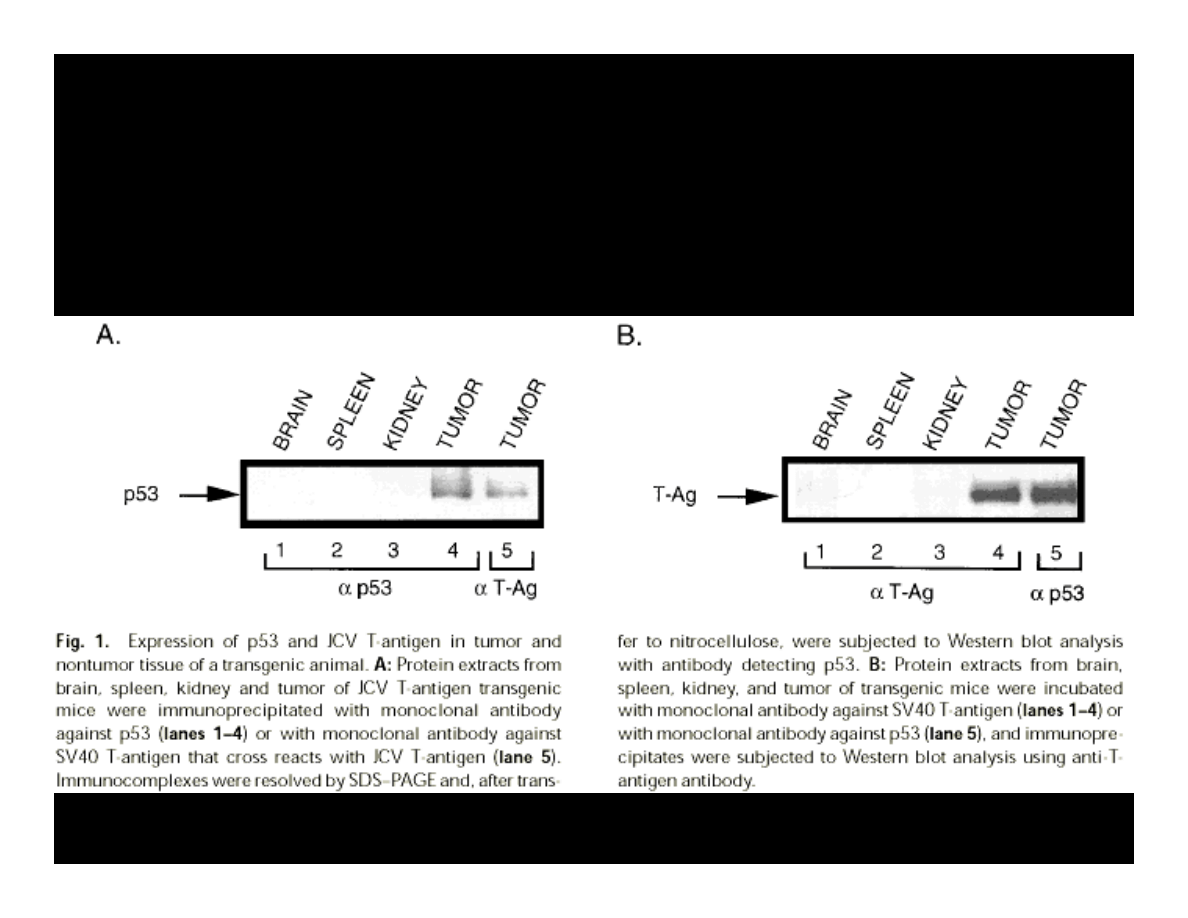
Krynska B., et al. (1997) JCB, 67:223-30
p21 EXPRESSION
IN THE PRESENCE OF T-ANTIGEN
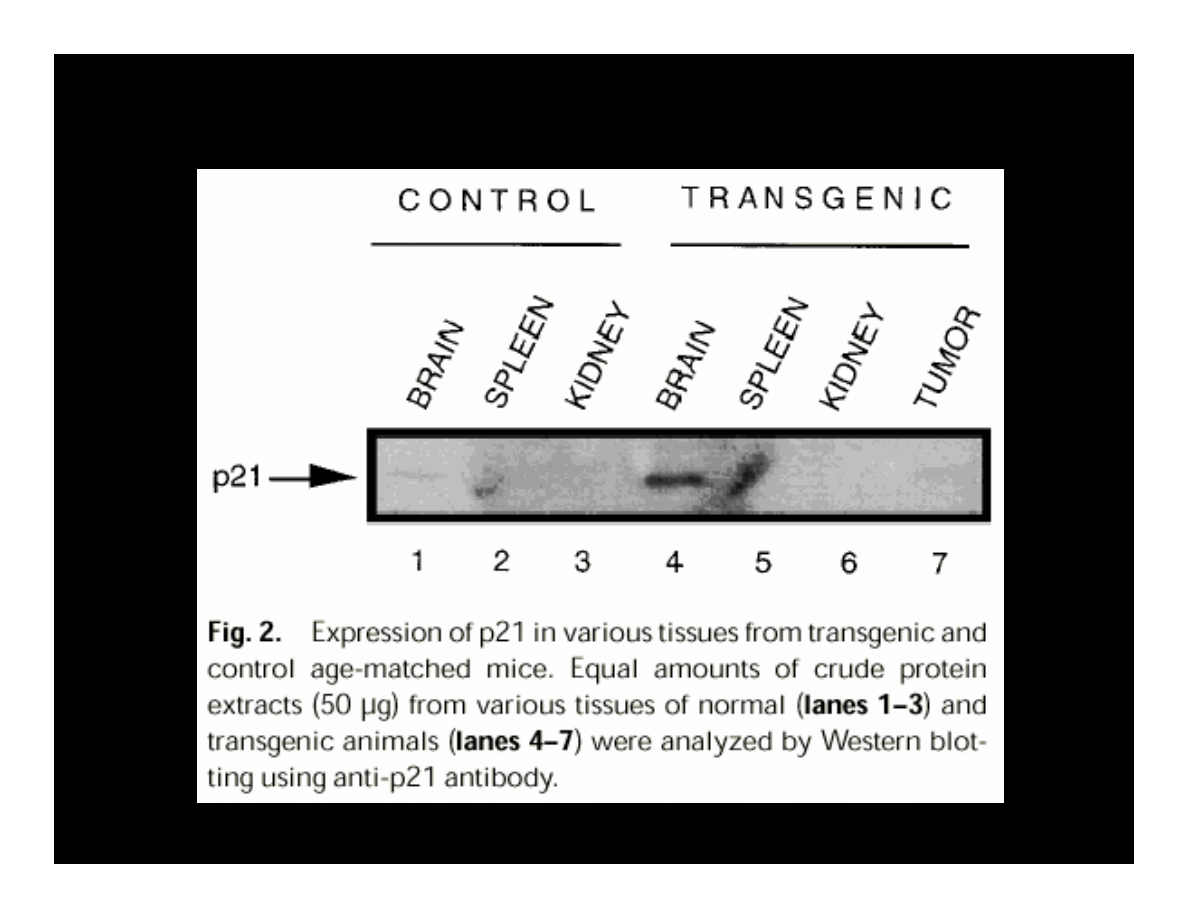
K
ry
ns
ka
B.
,
et
a
l.
(1
9
9
7
)
J
C
B,
6
7
:2
2
3
-3
0
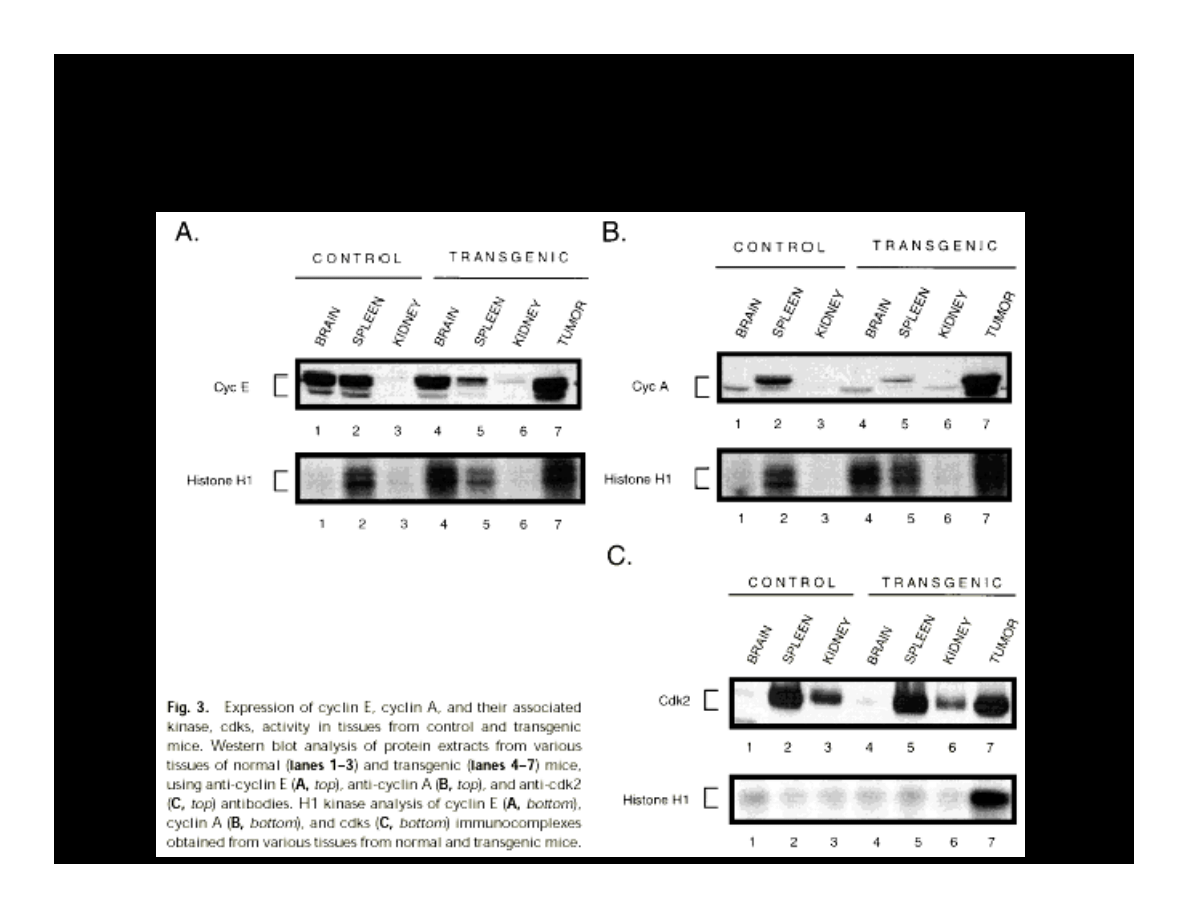
EXPRESSION OF CYCLIN E, A, & CDK2
IN THE PRESENCE OF T-ANTIGEN
K
ry
ns
ka
B.
,
et
a
l.
(1
9
9
7
)
J
C
B,
6
7
:2
2
3
-3
0
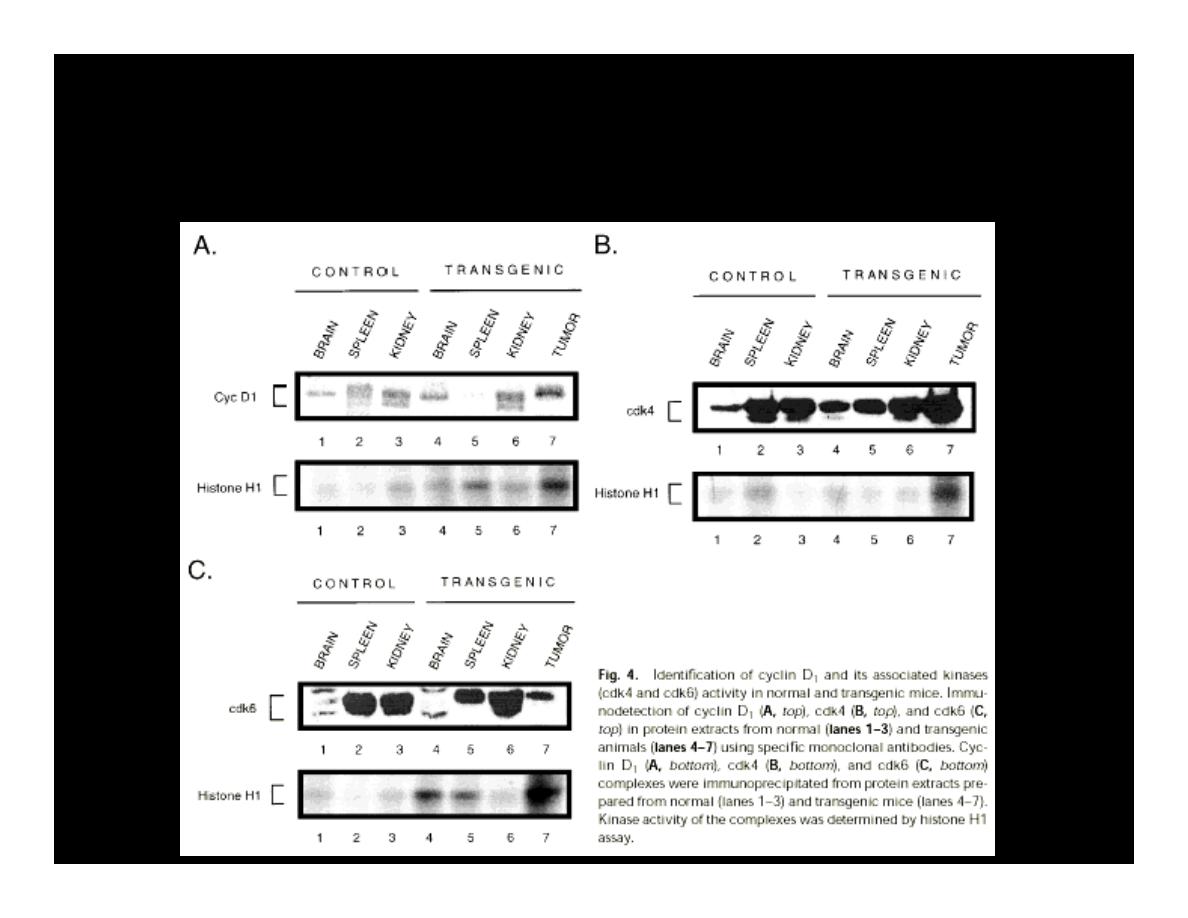
EXPRESSION OF CYCLIN D, CDK4, & CDK6
IN THE PRESENCE OF T-ANTIGEN
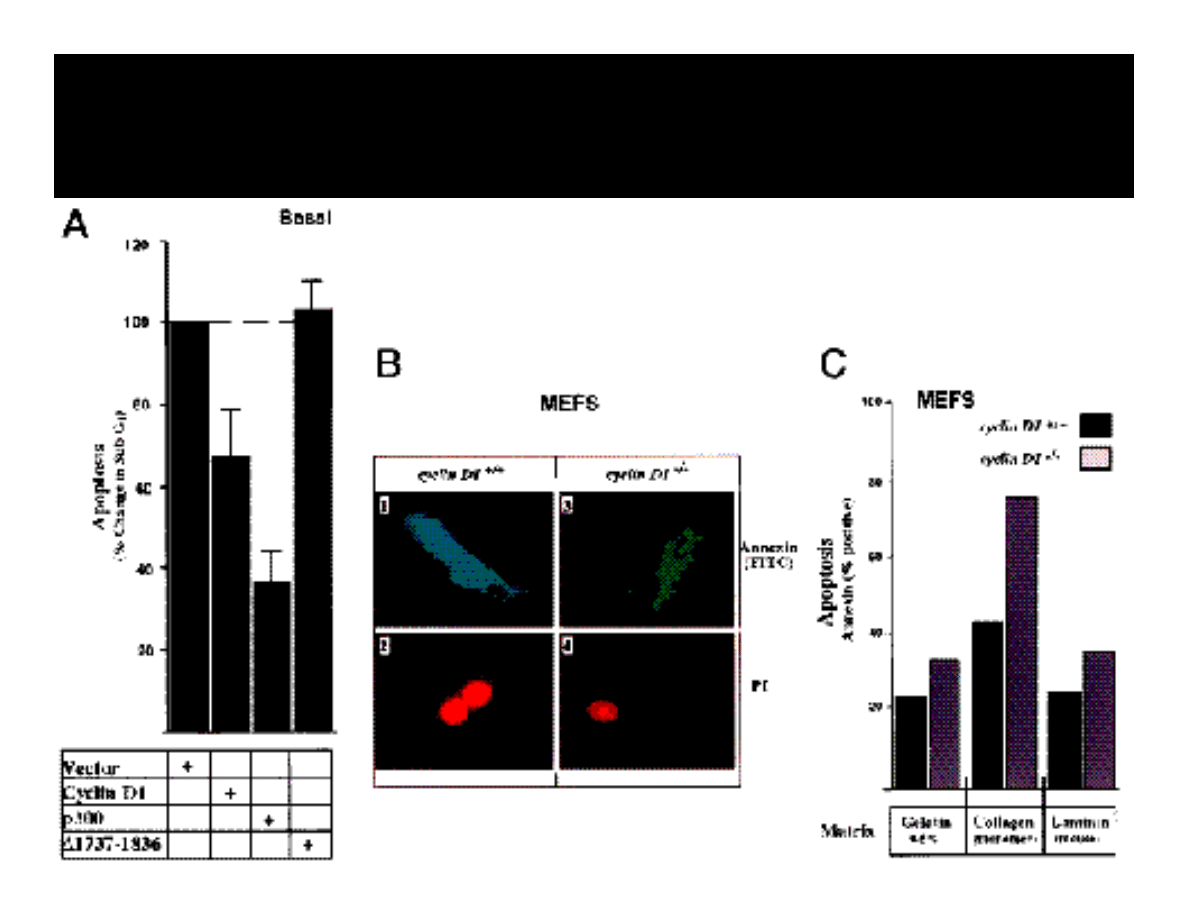
INHIBITION OF APOPTOSIS BY p300
REQUIRES PRESENCE OF CYCLIN D1
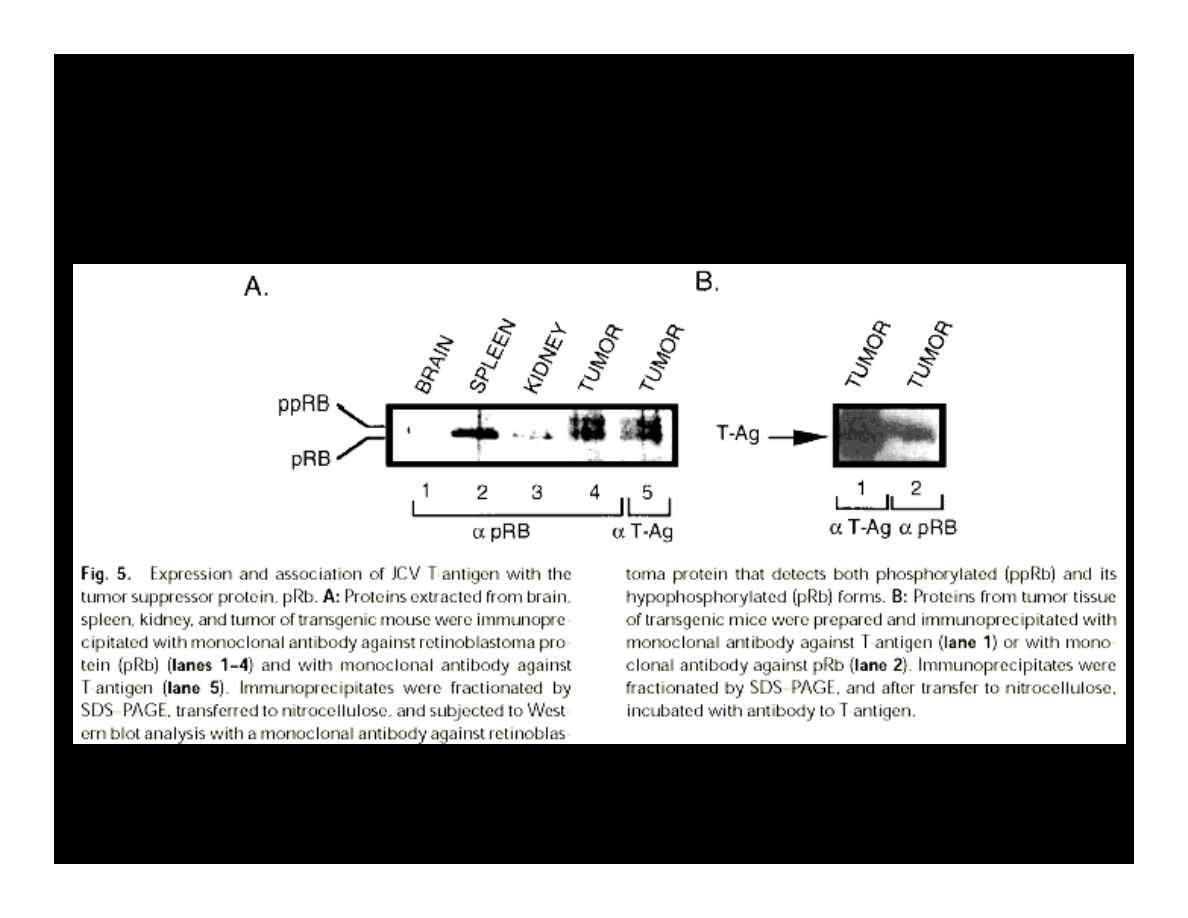
COMPLEXES OF T-ANTIGEN & pRB
Krynska B., et al. (1997) JCB, 67:223-30
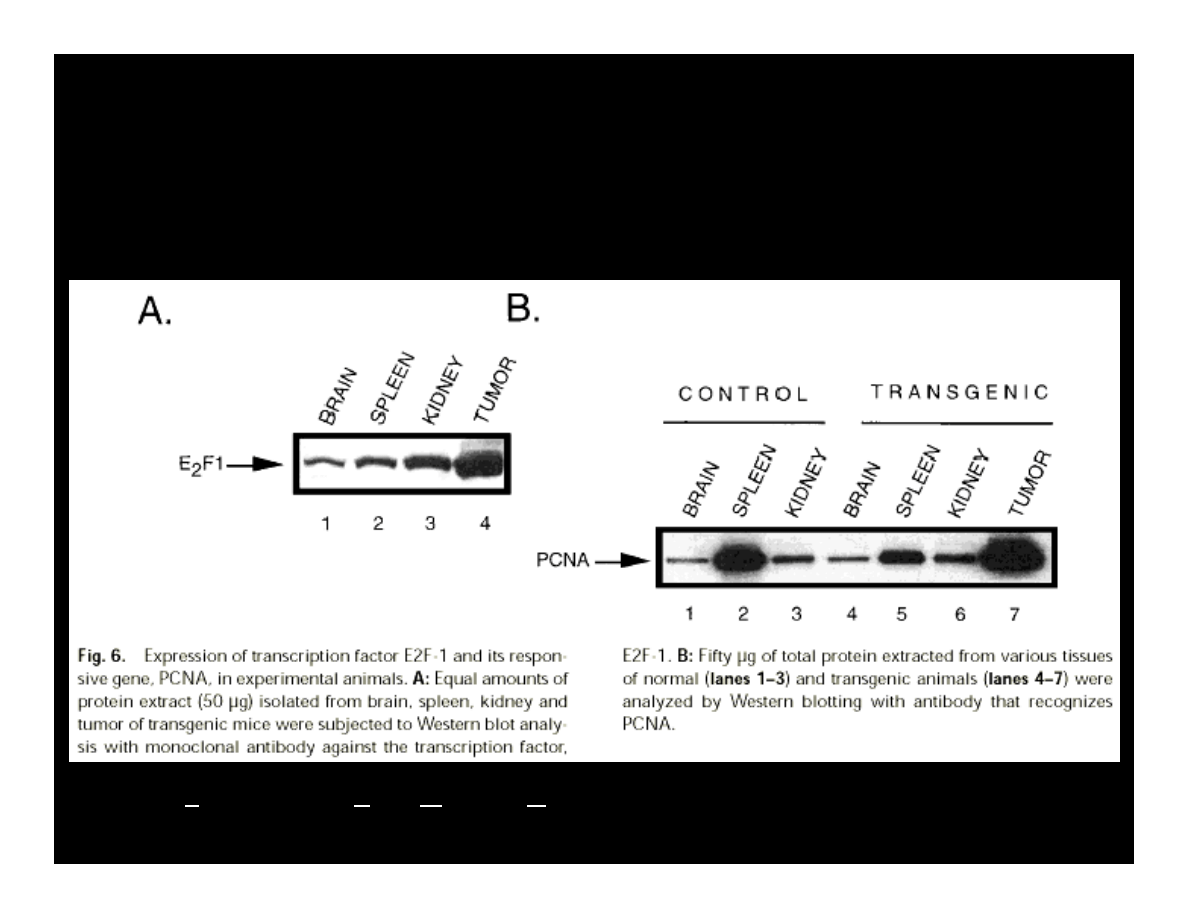
EXPRESSION OF E2F & PCNA
IN THE PRESENCE OF T-ANTIGEN
Krynska B., et al. (1997) JCB, 67:223-30
PCNA – Proliferation Cells Nuclear Antigen
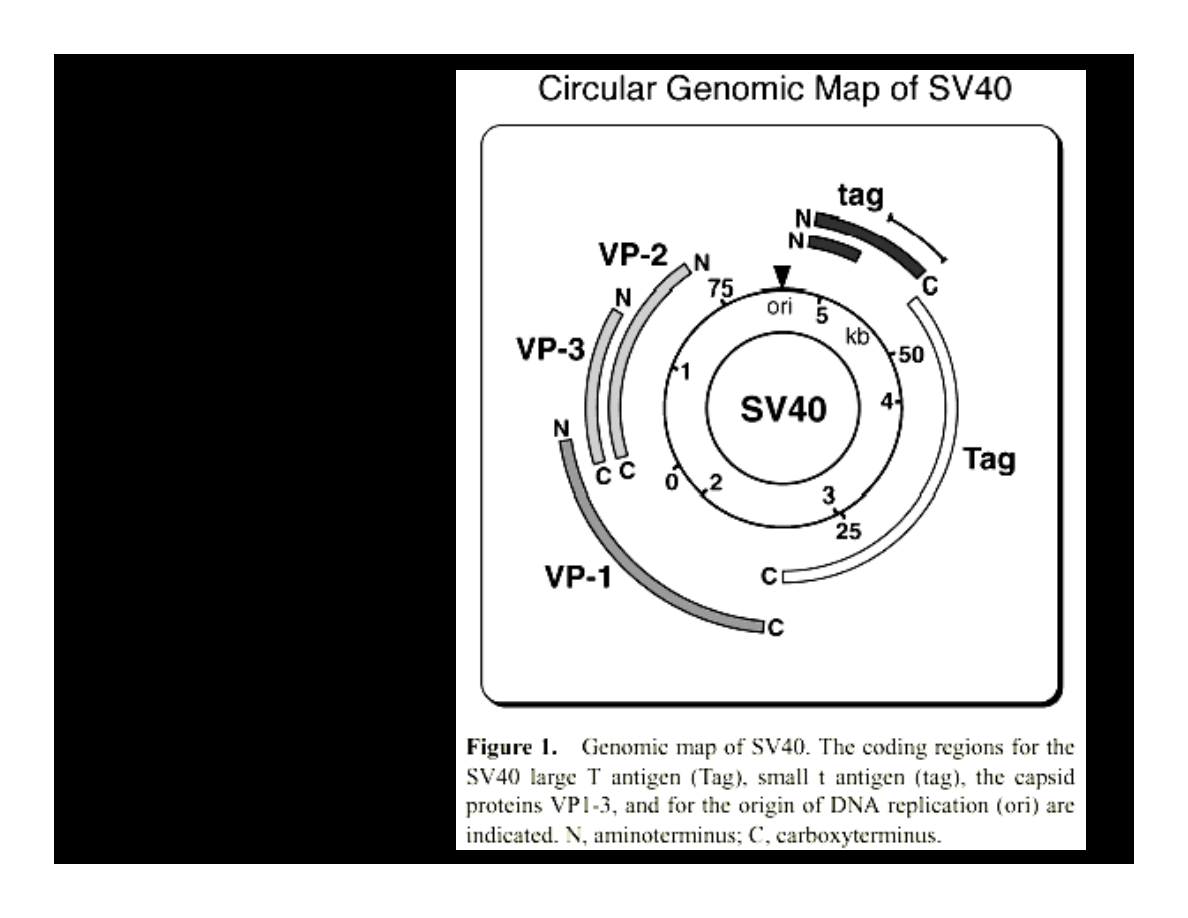
R
iz
zo
P.
,
et
a
l.
(2
0
0
1)
C
an
ce
r
Bi
ol
,
2
1:
6
3
-7
1
SV40
GENOME
PCR A M P L I F I C A T I O N
Immuno
cyto
SV40
JCV
Age
chemistry
T-Ag
T-Ag
T-Ag
No.
Sex
years
Diagnosis
T-Ag
(C-term)
(N-term)
(C-term)
VP-1
1
M
5
Classic
N/A
-
+
-
+
2
M
7
Desmoplastic
N/A
+
+
+
+
3
M
6
Desmoplastic
N/A
-
+
-
+
4
M
4
Classic
N/A
-
+
+
+
5
M
5
Desmoplastic
N/A
-
+
-
+
6
F
11
Classic
N/A
+
+
-
+
7
M
2
Neuroblastic
N/A
-
+
+
+
8
M
1.5
Desmoplastic
-
-
+
-
+
9
F
4
Desmoplastic
-
-
+
+
+
10
M
15
Desmoplastic
+
+
+
-
-
11
F
42
Desmoplastic
-
-
+
-
-
12
M
7
Classic
-
-
+
+
+
13
F
18
Neuroblastic
-
-
+
-
+
14
F
8
Desmoplastic
-
-
+
-
+
15
M
7
Classic
+
-
+
+
+
16
M
9
Classic
-
+
+
+
+
17
F
3
Desmoplastic
+
-
+
+
+
18
F
9
Desmoplastic
-
-
-
+
+
19
M
5
Neuroblastic
+
-
+
+
+
20
F
2
Classic
-
-
-
+
+
21
M
Newborn
Classic
-
-
+
+
+
22
M
12
Classic
-
+
-
-
-
23
M
5
Neuroblastic
-
-
+
+
+
Krynska B., et al.
(1999) PNAS,
96:11519-24
SV
40
&
JC
V
T-
AN
TI
GE
NS
PR
ES
EN
T
IN
AB
OU
T 2
2%
CA
SE
S

Table 2. SV40 DNA detection in human tumors: PCR based
evidence
Tumor type
Reference
Primer set (amplicon size bp)
Positivity
%
rate
Pleural
Griffiths
et al.
36
SV.for3/SV.rev (105)
26/26
100
mesothelioma
SV.for2/SV.rev (574)
3/26
12
De Luca
et al.
27
SV.for3/SV.rev (105)
30/35
86
Pass
et al.
30
SV.for3/SV.rev (105)
30/42
71
Mayall
et al.
41
SV.for3/SV.rev (105)
5/7
71
Shivapurkar
et al.
44 SV.for3/SV.rev (105)
61/93
66
Carbone
et al.
16
SV.for3/SV.rev (105)
29/48
60
PYV.for/PYV.rev (172)
36/48
75
Procopio
et al.
54
PYV.for/PYV.rev (172)
50/83
60
Ramael
et al.
103
PYV.for/PYV.rev (172)
14/25
56
SV.for2/SV.rev (574)
Cristaudo
et al.
48
RA3/RA4 (482)
10/18
56
Galateau-Salle
PYV.for/PYV.rev (172)
10/21
48
et al.
33

Table 2. SV40 DNA detection in human tumors: PCR based
evidence (cont. 1)
Tumor type
Reference
Primer set (amplicon size bp)
Positivity
%
rate
Pleural
Dhaene
et al.
40
SV.for3/SV.rev (105)
13/28
46
mesothelioma
Pepper
et al.
18
SV.for3/SV.rev (105)
4/9
44
PYV.for/SV.rev (172)
6/9
67
Strizzi
et al.
47
PYV.for/PYV.rev (172)
9/23
39
Mutti
et al.
100
SV.for3/SV.rev (105)
8/25
32
Strickler and
SV.for3/SV.rev (105)
0/50
0
Goedert 23
Mulaterro
et al.
63
PYV.for/SV.rev (172)
0/12
0
Hirvonen
et al.
37
SV2for/SV.rev (576)
0/49
0
Emri
et al.
51
SV.for2/SV.rev (574)
0/29
0
Table 2. SV40 DNA detection in human tumors: PCR based
evidence (cont. 2)
Tumor type
Reference
Primer set (amplicon size bp)
Positivity
%
rate
Peritoneal
Shivapurkar
et al.
44 SV.for3/SV.rev (105)
6/11
55
mesotheliomas
Brochopulmonary
Galateau-Salle
PYV.for/PYV.rev (172)
18/63
29
carcinomas
et al.
33
Shivapurkar
et al.
44 SV.for3/SV.rev (105)
0/20
0
Procopio
et al.
34
PYV.for/SV.rev (172)
0/18
0
Carbone
et al.
16
SV.for3/SV.rev (105)
0/13
0
Pepper
et al.
18
SV.for3/SV.rev (105)
0/9
0
SV.for3/SV.rev (105)
Choroid plexus
Martini
et al.
20
PYV.for/PYV.rev (172)
5/6
83
tumours
Bergsagel
et al.
14
SV.for3/SV.rev (105)
10/20
50
Huang
et al.
39
SVTAGP1/SVTAGP3 (158)
6/16
38
Lednicky
et al.
106
LA1/LA2 (294)
10/13
77
Bergsagel
et al.
14
SV.for3/SV.rev (105)
10/20
50
Ependymomas
Bergsagel
et al.
14
SV.for3/SV.rev (105)
10/11
91
Lednicky
et al.
106
LA1/LA2 (294)
3/3
100
Martini
et al.
20
PYV.for/PYV.rev (172)
8/11
73

Table 2. SV40 DNA detection in human tumors: PCR based
evidence (cont. 3)
Tumor type
Reference
Primer set
Positivity
%
(amplicon size bp)
rate
Ependymomas
Weggen
et al.
53
SV.for3/SV.rev (105)
1/25
4
Bersagel
et al.
14
SV.for3/SV.rev (105)
7/11
64
Medulloblastomas
Lednicky
et al.
106
LA1/LA2 (294)
2/6
33
Huang
et al.
39
SVTAGP1/SVTAGP3 (158)
5/17
29
Weggen
et al.
53
SV.for3/SV.rev (105)
2/116
2
Krynska
et al.
102
SV.for3/SV.rev (105)
5/23
22
Astrocytomas
Martini
et al.
20
PYV.for/PYV.rev (172)
8/17
17
Huang
et al.
39
SVTAGP1/SVTAGP3 (158)
22/50
44
Meningiomas
Martini
et al.
20
PYV.for/PYV.rev (172)
1/7
14
Weggen
et al.
53
SV.for3/SV.rev (105)
1/131
1
Oligodendroglioma
Huang
et al.
39
SVTAGP1/SVTAGP3 (158)
3/12
25
Martini
et al.
20
PYV.for/PYV.rev (172)
0/9
0
Glioblastomas
Martini
et al.
52
PYV.for/PYV.rev (172)
10/30
33

Table 2. SV40 DNA detection in human tumors: PCR based
evidence (cont. 4)
Tumor type
Reference
Primer set
Positivity
%
(amplicon size bp)
rate
Osteosarcomas
Lednicky
et al.
15 SV.for3/SV.rev (105)
5/10
50
TA1/TA2 (441)
5/10
50
Yamamoto
et al.
50 R1/R2
(315)
21/54
39
Carbone
et al.
17
SV.for2/SV.rev (574)
40/126
32
Mendoza
et al.
32 SV.for3/SV.rev (105)
9/35
26
Other bone
Carbone
et al.
17
SV.for2/SV.rev (574)
14/34
32
Tumours
Gamberi
et al.
46 PYV.for/SV.rev (172)
30/107
28
SV.for2/SV.rev (574)
Papillary thyroid Pacini
et al.
31
SV.for3/SV.rev (105)
3/69
4
carcinomas (PTC)
LA1/LA2 (294)
Non-PTC thyroid Pacini
et al.
31
SV.for3/SV.rev (105)
0/9
0
tumours
LA1/LA2 (294)
Non-Hodgkin’s
Rizzo
et al.
38
V5/SV6 (172)
3/29
10
lymphomas
Martini
et al.
52
PYV.for/PYV.rev (172)
13/95
14

Table 2. SV40 DNA detection in human tumors: PCR based
evidence (cont. 5)
Tumor type
Reference
Primer set
Positivity
%
(amplicon size bp)
rate
Hodgkin’s
Martini
et al.
52
PYV.for/PYV.rev(172)
8/55
15
lymphomas
AIDS lymphoma
Rizzo
et al.
38
SV5/SV6 (172)
2/5
40
Prostatic tumours Radu
et al.
68
SV.for3/SV.rev (105)
4/33
12

2948–2953, PNAS, March 5, 2002, vol. 99, no. 5
Targeted point mutations of p53 lead to
dominant-negative inhibition of wild-type
p53 function
Annemieke de Vries* †‡ , Elsa R.
Flores*, Barbara Miranda* † , Harn-Mei
Hsieh*, Conny Th. M. van Oostrom ‡ ,
Julien Sage*, and Tyler Jacks*
† Howard Hughes Medical Institute,
Massachusetts Institute of Technology,
Cambridge, MA
and ‡ National Institute of Public Health,
Laboratory of Health Effects Research, 3720 BA
Bilthoven, The Netherlands
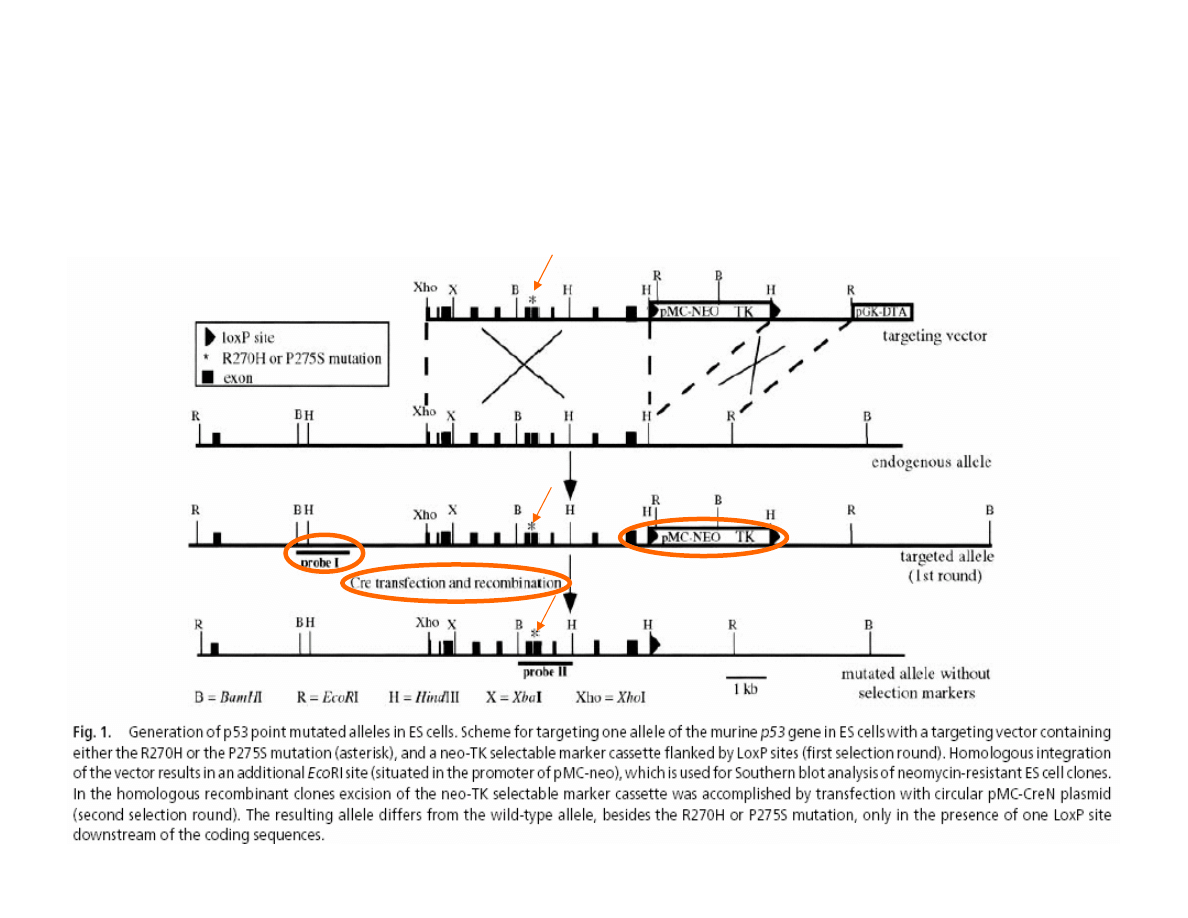
Mutation
Mutation
Mutation
Homologous recombination
ONE ALLELE MUTATION OF p53 GENE
de Vries A. et al. (2002) PNAS, 99:2948-53.
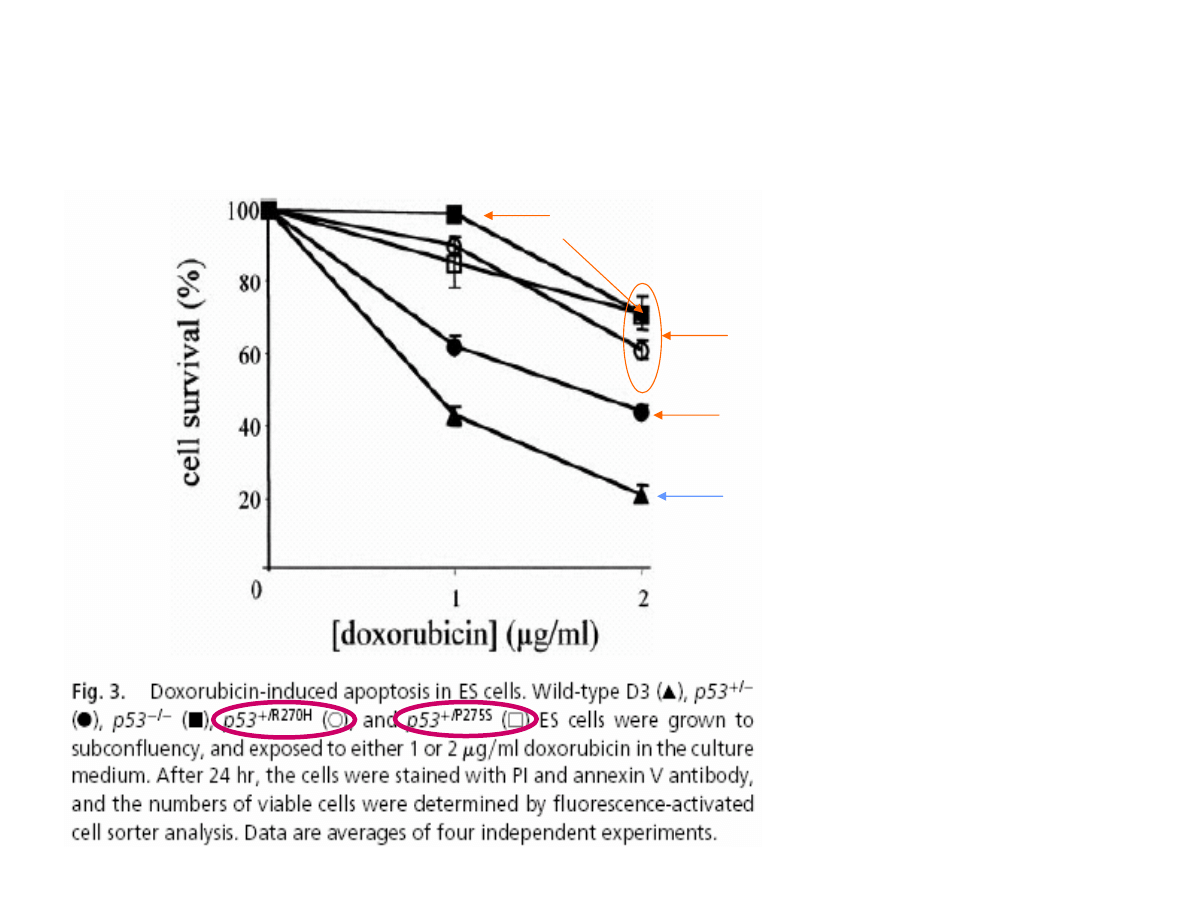
EFFECT OF ONE ALLELE MUTATION OF p53 GENE
ON EMBRYONIC CELLS SURVIVAL
NORMAL/
WILDE p53 GENE
ONE p53 ALLELE
INACTIVE
HETEROZYGOUS
p53 MUTATION
de Vries A. et al.
(2002) PNAS, 99:2948-53.
TWO p53 ALLELES
INACTIVE
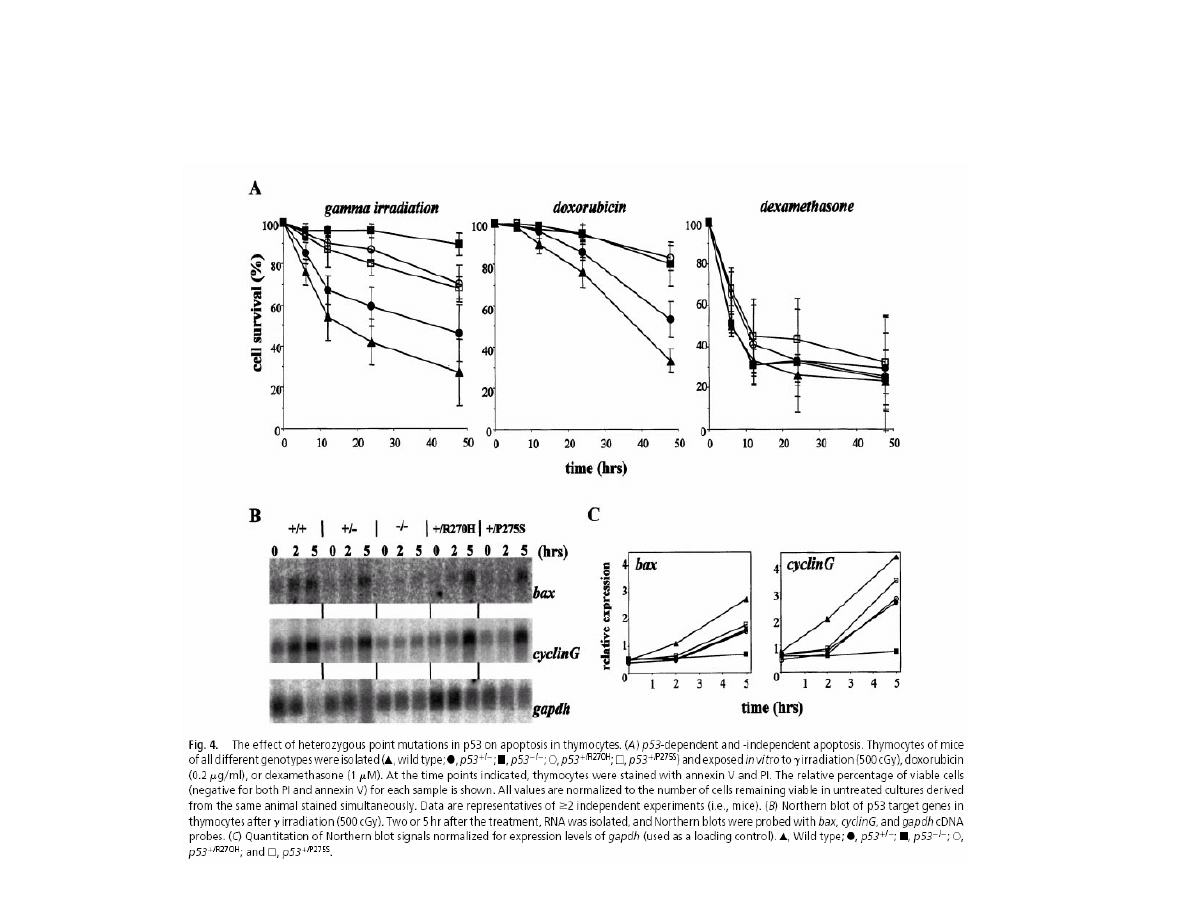
p53 DEPENDENT APOPTOSIS
p53 INDEPENDENT
APOPTOSIS
EFFECT OF ONE ALLELE p53 GENE MUTATION
ON THYMUS CELLS SURVIVAL
d
e
V
ri
e
s
A
.
e
t
a
l.
(2
0
0
2
)
PN
A
S
,
9
9
:2
9
4
8
-
5
3
.

1–5, Nature Genetics, FEBRUARY 11, 2002, online publication
BRCA1 regulates the G2/M checkpoint by
activating Chk1 kinase upon DNA damage
Ronit I. Yarden
1
, Sherly Prado-Reoyo
1
,
Magda Sgagias
2
, Kenneth H. Cowan
2
&
Lowrence C. Brody
1
1 Genome Technology Branch, National Human
Genome Research Institute, National Institutes
of Health, Bethesda, MD
2 Eppley Institute of Cancer Research,
University of Nebraska Medical Center, NA
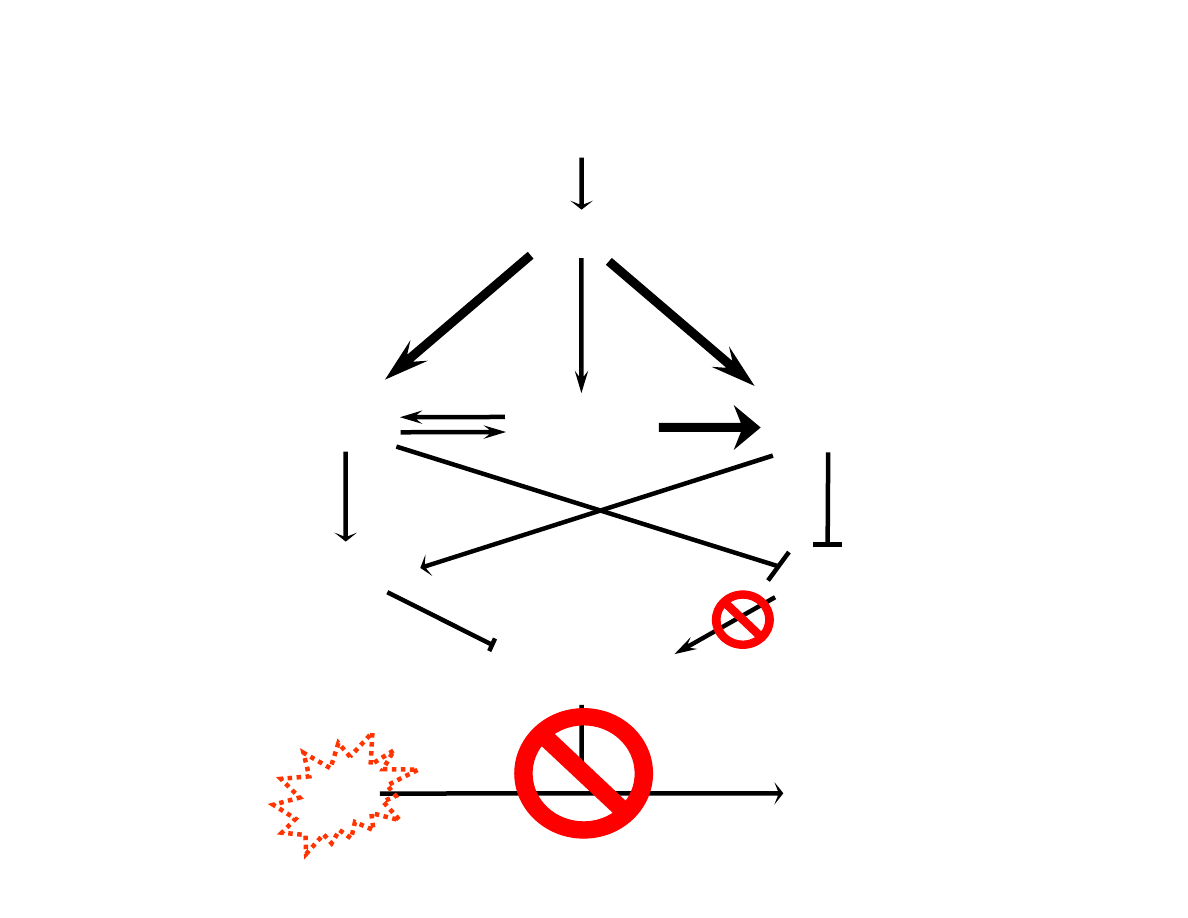
DNA Damage
ATM/ATR
Chk1 –
A Regulatory Kinase
hCds1/Chk2
?
Cdc25C
Wee1
Kinase
Cdc2/cyclin B
G2
M
ATM, ATR and hCds1/Chk2
Are DNA Damage Response
Proteins Changing
BRCA phosphorylation
BRCA1
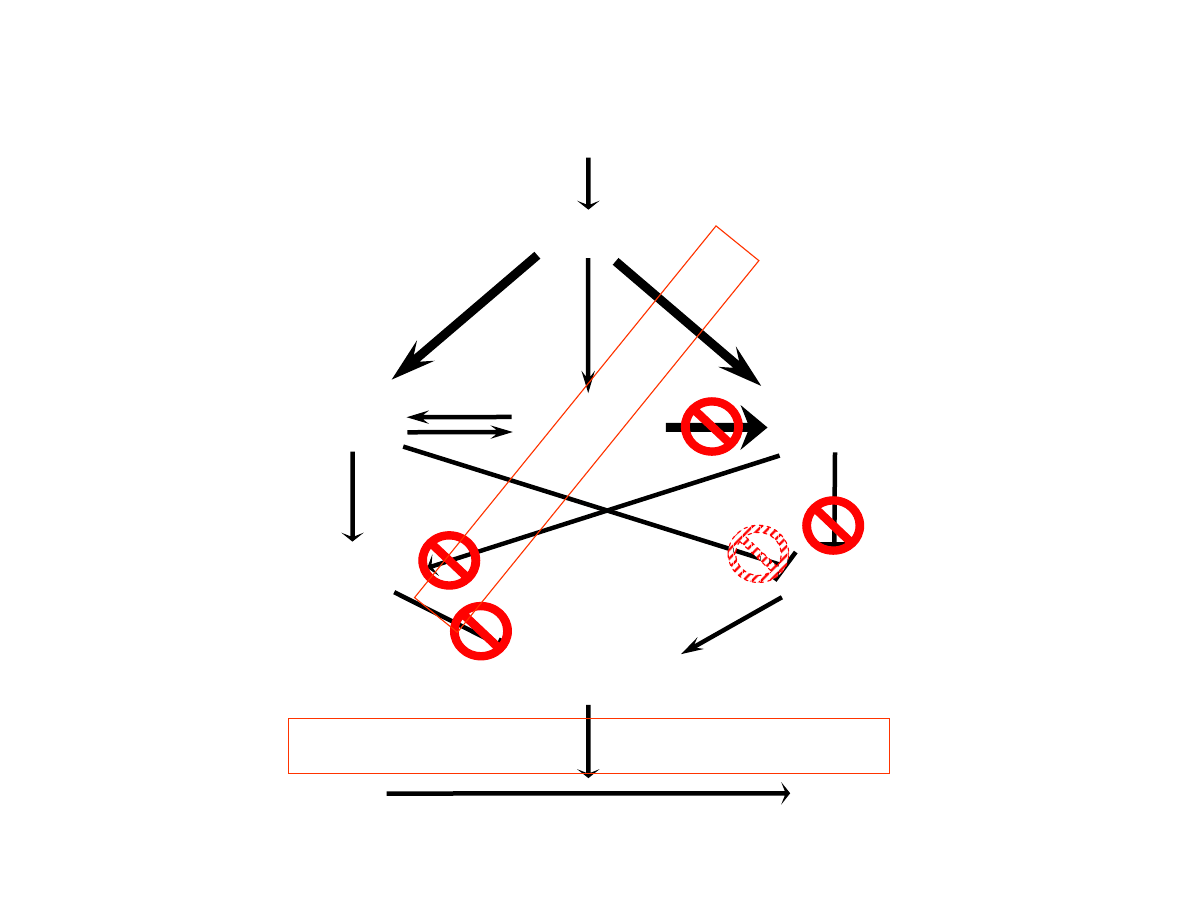
DNA Damage
ATM/ATR
BRCA1
Chk1 -
A Regulatory Kinase
hCds1/Chk2
?
Cdc25C
Wee1
Kinase
Cdc2/cyclin B
G2
M
ATM, ATR and hCds1/Chk2
Are DNA Damage Response
Proteins Changing
BRCA phosphorylation
UNCONTROLED PROLIFERATION
LA
C
K
O
F
B
R
C
A
1
A
C
T
IV
IT
Y
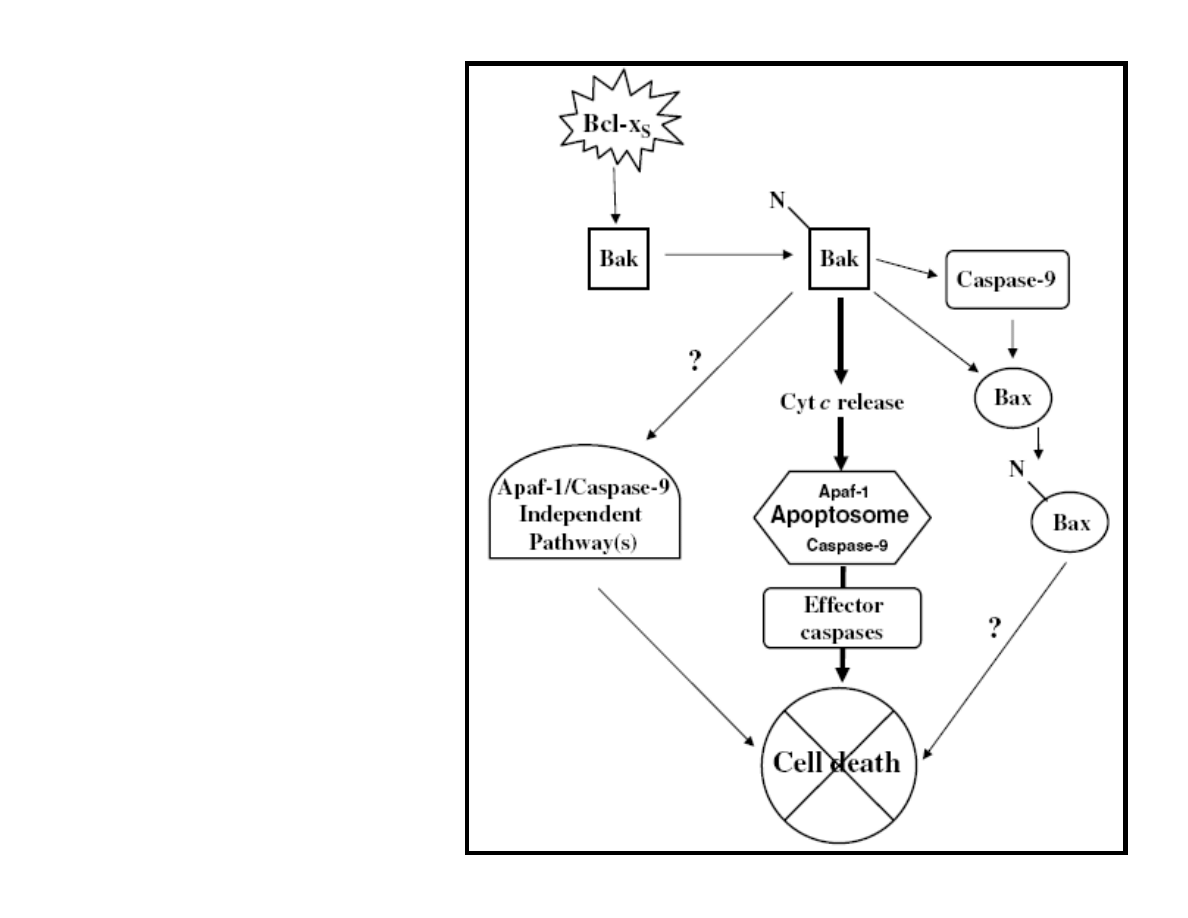
Lindenboim L., et al. Cell Death and
Differentiation (2005) 12, 713–723
Figure 6 A model depicting the
apoptotic effects of Bcl-xS in MEFs.
Expression
of Bcl-xS in MEFs induces exposure
of NT of Bak, leading to its
activation. The
activated Bak can induce three
pathways: a major pathway leading to
cytochrome c release, which in turn
causes apoptosome activation and cell
death in a caspase-dependent manner;
a second pathway, which leads to
Apaf-
1- and caspase-9-independent cell
death; and a third pathway, which
induces the
exposure of Bax NT, both in a
caspase-9-dependent and -
independent manner,
causing its activation. The first and
the second pathways contribute to the
death
process. The role of the third pathway
is not yet known.
Uncontroled divisions
Early Changes
Late Changes
CANCER
Cancer In Situ
Malignant Form (Invasive)
Normal Epithelium
Precancer
Metastasis
Progresive Cumulation
of Gene Damage
CANCER TRANSFORMATION
LOS
S O
F SO
CIA
L CO
NTR
OL
(APO
PTO
SIS
)

DNA repair disorders may
complicate,
if the disorder causes the patient to be unusually
sensitive to the therapy:
1. cancer chemotherapy
&
2. radiation therapy
MAJOR CONCLUSION
Cancer genetics
Cancer genetics

THANK YOU
FOR YOUR ATTENTION
Wyszukiwarka
Podobne podstrony:
Niestabilność genetyczna w nowotworach 1
genetyka nowotworow
genetyka nowotworow
Genetyka nowotworów
Niestabilność genetyczna w nowotworach., Niestabilność chromosomowa:
03 Genetyka w nowotworachid 4362 ppt
Genetyka nowotworów
Niestabilność genetyczna w nowotworach, NIESTABILNOŚĆ GENETYCZNA W NOWOTWORACH
14 Genetyka nowotworow
Niestabilność genetyczna w nowotworach 1
Genetyka nowotworzenia 01
Genetyka nowotworów
Genetyczne podstawy nowotworów, Biologia medyczna
Diagnostyka predyspozycji genetycznych do chorob nowotworowych
GENETYCZNE PODSTAWY NOWOTWORÓW(1)
GENETYKA KLINICZNA V rok seminarium Nowotwory dziedziczne wprowadzenie Nowotwory jelita grubeg
nowotwory dla studentów, psychologia, Genetyka
IG.4 - Uszkodzenia i naprawa DNA w komórkach nowotworowych, Genetyka, Inżynieria genetyczna
więcej podobnych podstron