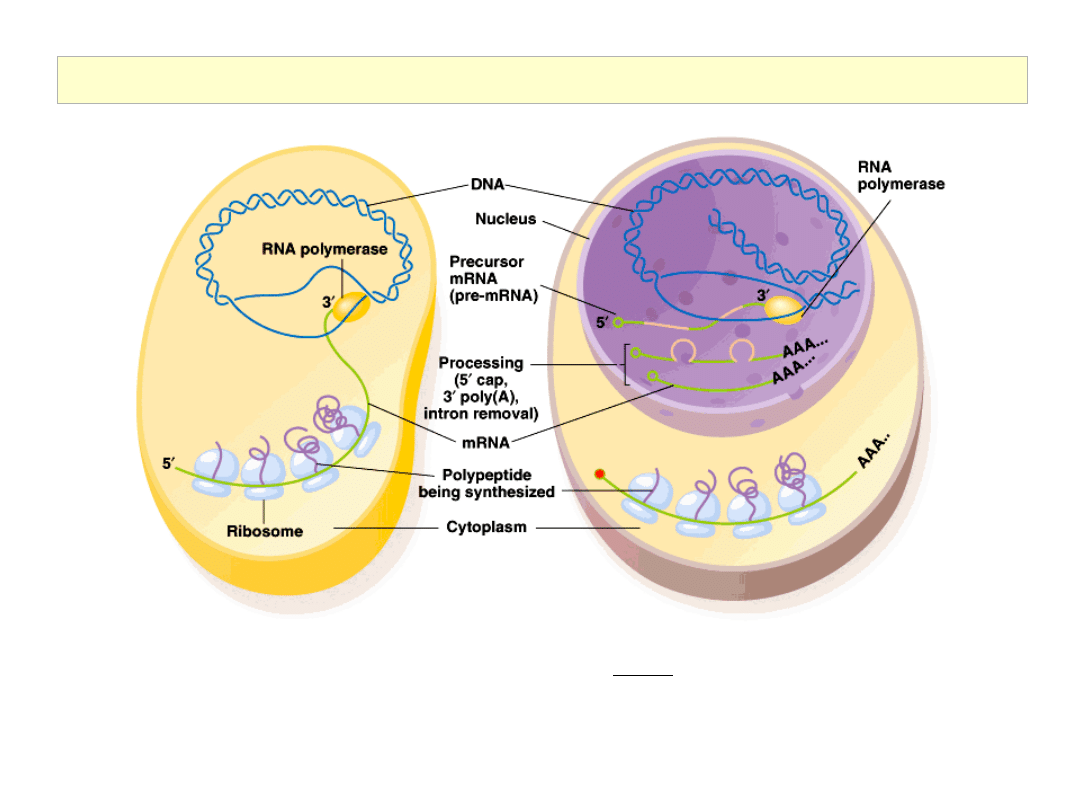
The Initiation of Translation in Pro- and Eukaryotic Cells
5’ cap:
After 20-30 nucleotides have been
synthesized, the 5’-end of the mRNA is capped
5’ to 5’ with a guanine nucleotide. Essential for
the ribosome to bind to the 5’ end of the mRNA.
Poly (A) tail:
50-250 adenine nucleotides are added to
3’ end of mRNA. Stabilizes the mRNA, and plays
an important role in transcription termination.
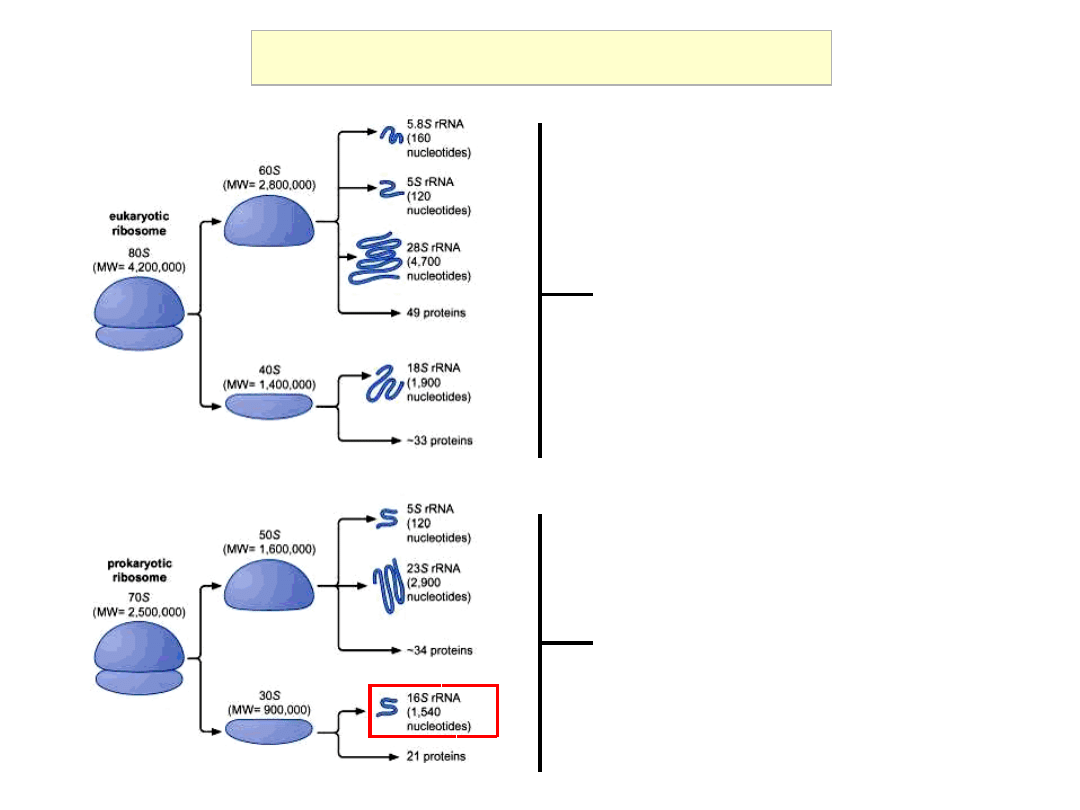
Eu- and Prokaryotic Ribosomes
Eukaryotic cytoplasm
Prokaryotes, Eukaryotic organelles
(mitochondria, chloroplasts)
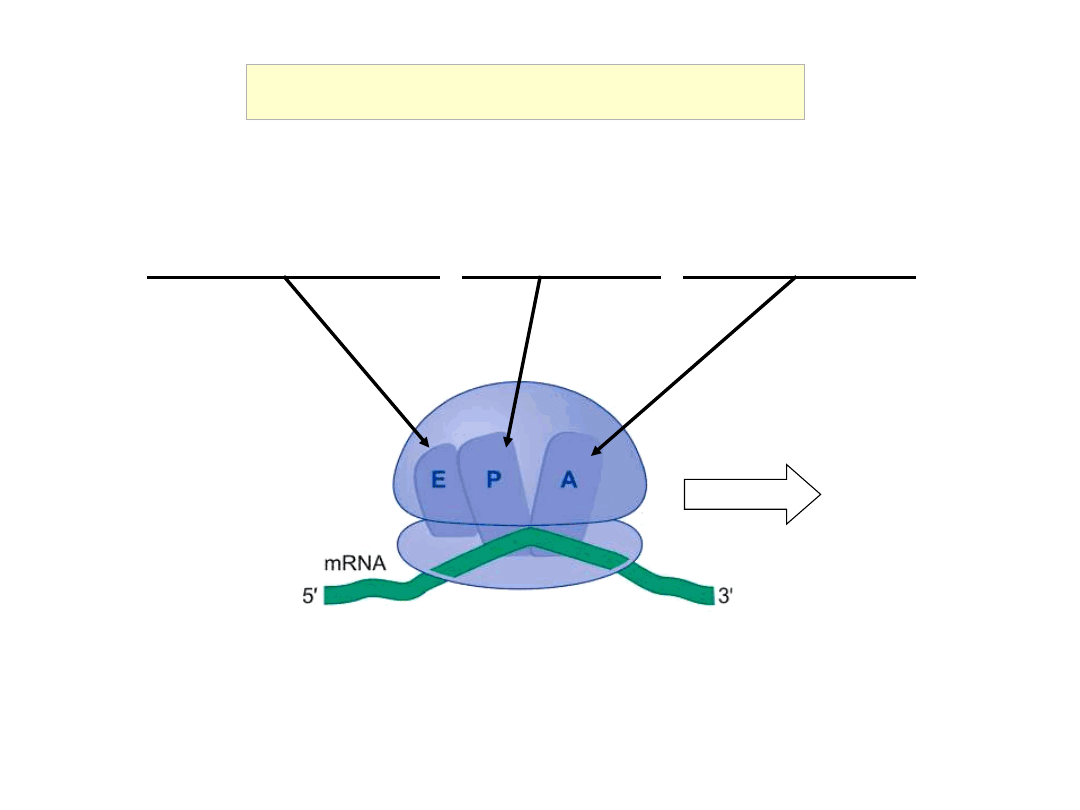
E: Exit site for free tRNA P: peptidyl-tRNA A: aminoacyl-
tRNA
E, P and A Sites of Ribosomes
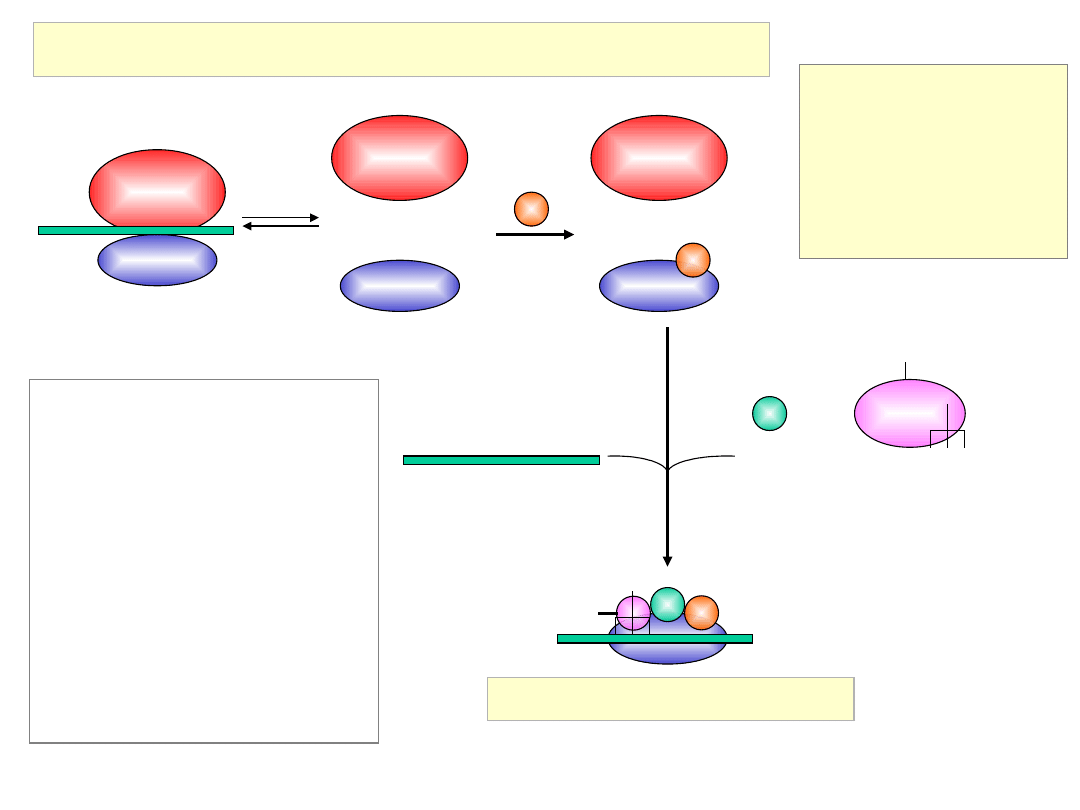
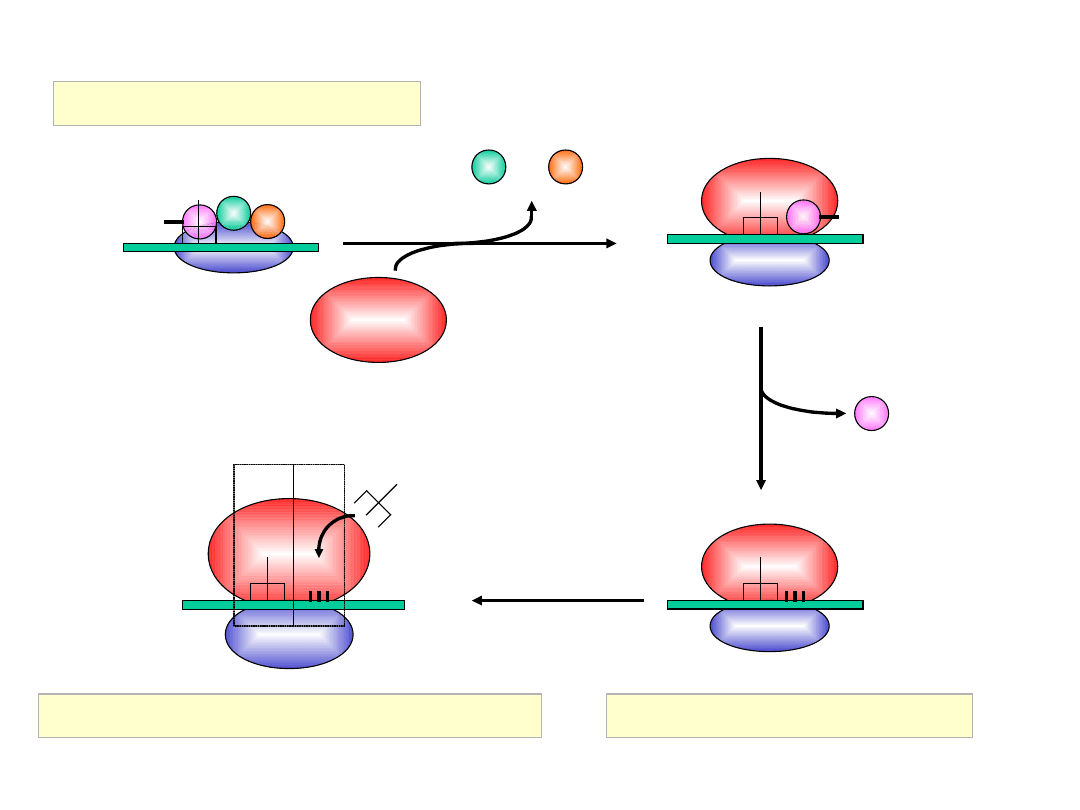
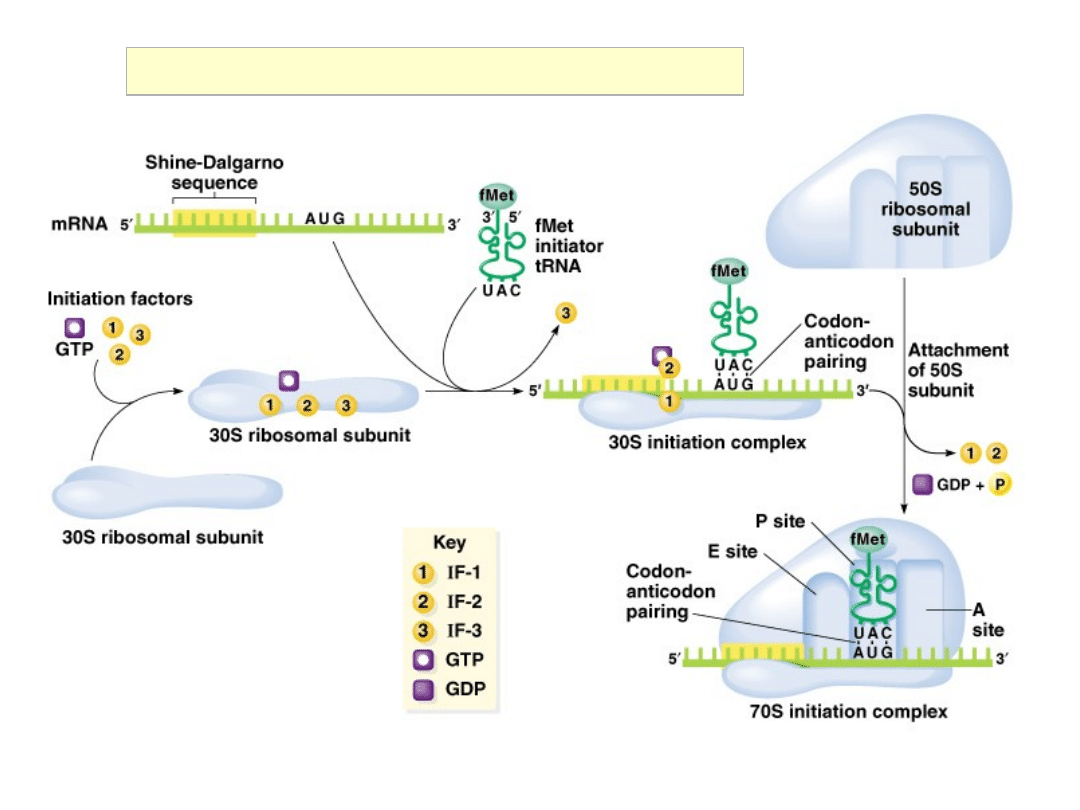
Initiation of Translation in Prokaryotes
50S
30S
50S
30S
+
RRF
+
3
IF-3
3
1
+
mRNA
(fMet-tRNA
f
Met
)
IF-2GTPfMet-tRNA
f
met
3
1
2
GTP
fMet
30S Initiation Complex
IF-1: 71aa, assists IF-2 binding
IF-2: 890aa, binds initiator
tRNA
and GTP
IF-3: 180aa, releases mRNA
and
tRNA from recycled 30S
subunit and aids (new)
mRNA binding
RRF: ribosome release factor
IF-2 = initiation factor 2
In complex with GTP, it brings
fMet-tRNA
f
Met
to the partial P
site on the small subunit.
Activates a GTPase activity in
the small subunit, which
allows dissociation of IF2, IF3,
and IF1.
mRNA
2
GTP
fMet
3
1
2
GTP
fMet
30S Initiation Complex
1
3
+
2
GTP
fMet
2
+ GDP +Pi
fMet
70S Initiation Complex
fMet
a
a
A
site
P
site
Elongation Phase of Translation
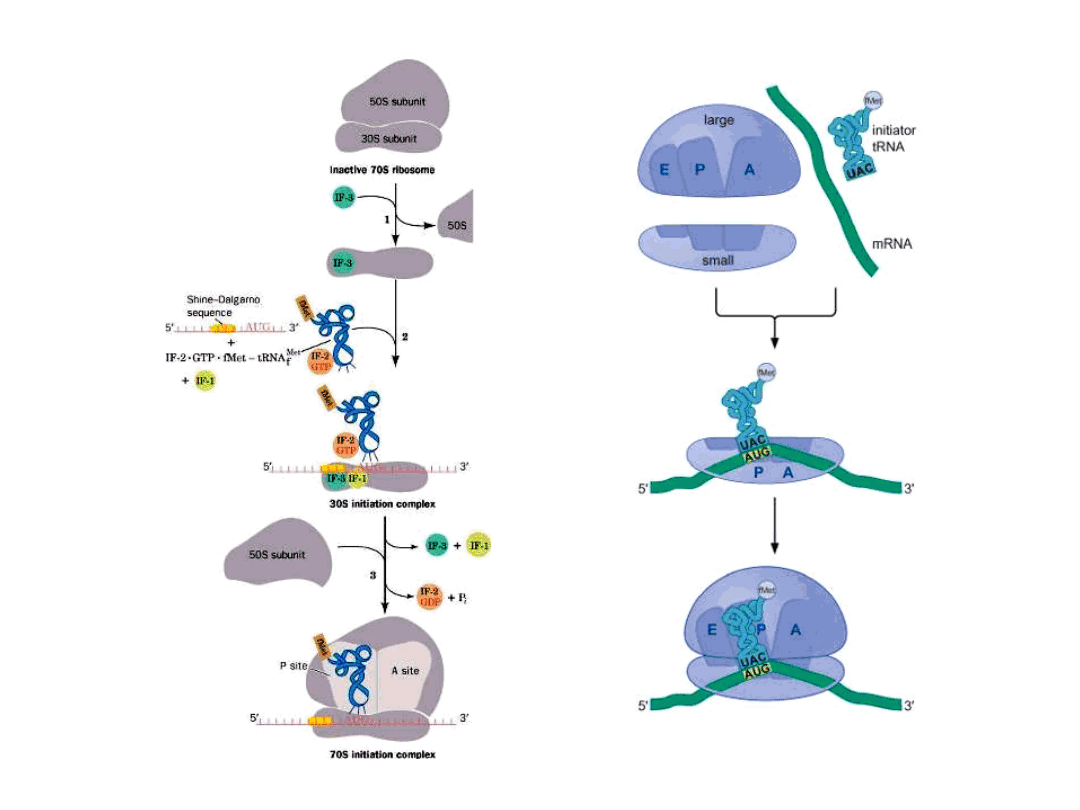
Simple process – involves only initiation factors (IFs) IF-1, IF-2
and IF-3
plus….. fMet-tRNA
f
Met
and mRNA
mRNA binds to small ribosomal subunit such that initiator AUG
is positioned in the precursor to the P site
In eubacteria, such as E. coli, the positioning of the initiator
AUG is mediated by base pairing between
the ribosome-binding site in the (5’) untranslated region of the
mRNA
and the 3’ end of the 16S rRNA
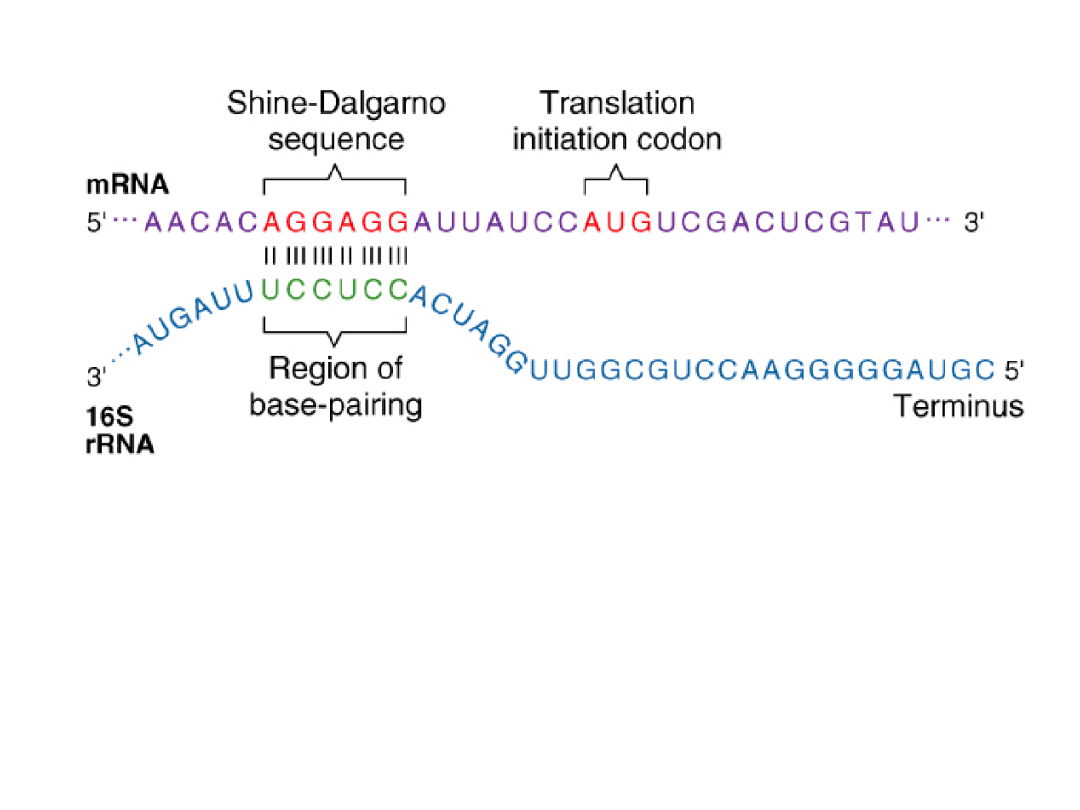
Initiation of Translation in Prokaryotes
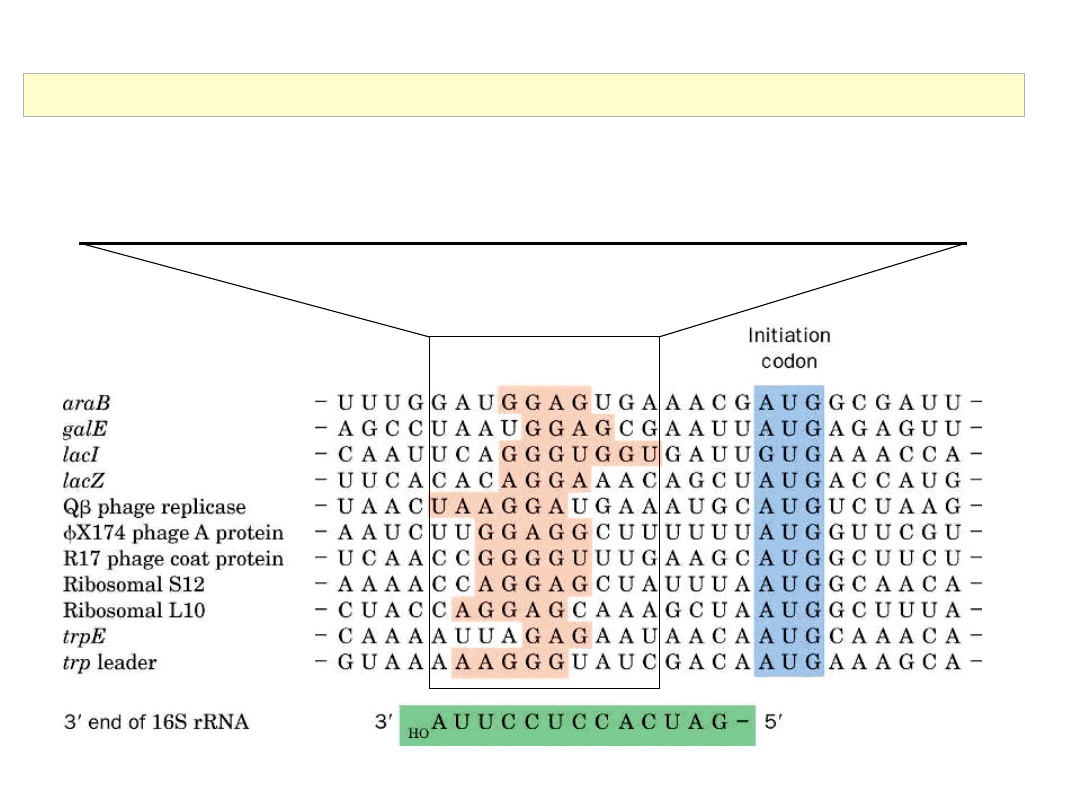
Some translational initiation sequences recognized by E. coli ribosomes.
Shine-Dalgarno (ribosome binding) sequence: A nucleotide sequence (consensus = AGGAGG)
that is present in the (5') untranslated region(s) of prokaryotic mRNAs. This sequence serves as a
binding site for ribosomes.
No involvement of mRNA 5’ end
Shine – Dalgarno sequences +AUG initiation codons can occur
within 5’ non-translated regions, and, may also occur within
site(s) internal to the mRNA …….
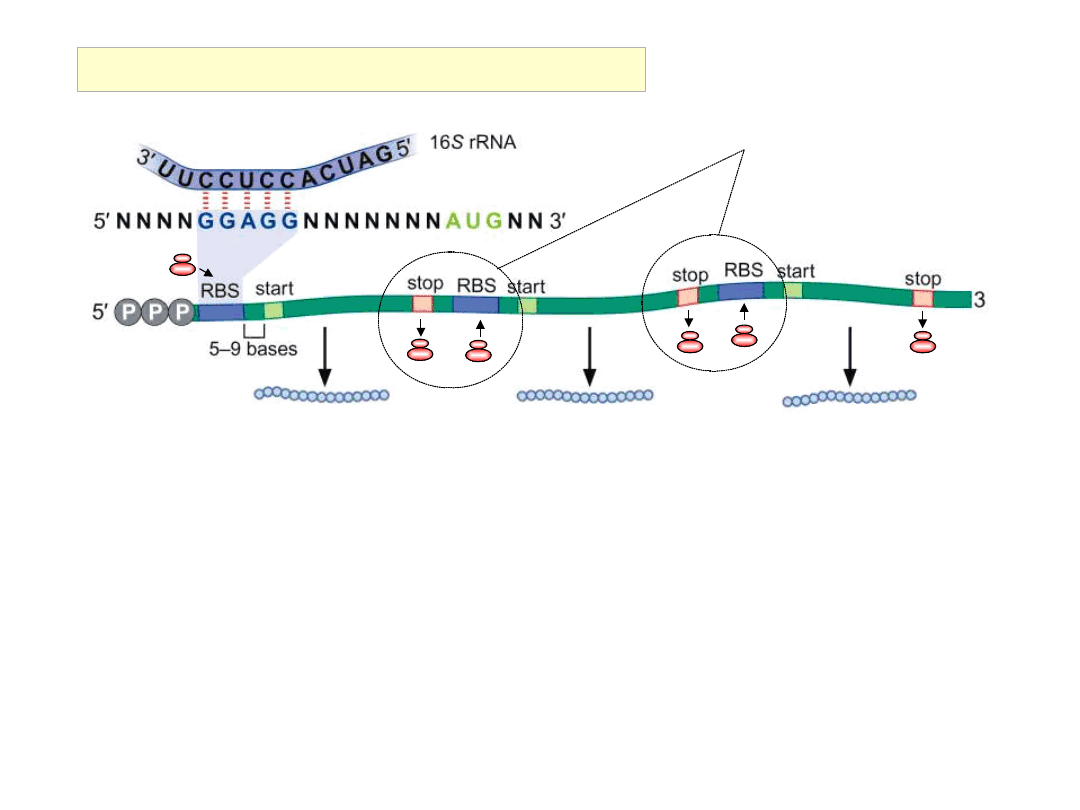
Prokaryotic mRNAs may be polycistronic
The ability to bind ribosomes and initiate translation at sites
internal to the prokaryotic mRNA allows
• genes to be organised into operons,
• an operon to be transcribed into a single (polycistronic) mRNA,
• the expression of a number of genes (related functions) to be
controlled
by a single promoter (or single translational control
mechanism)
cistron 1
cistron 2
cistron 3
sites of ribosome ‘re-cycling’
Initiation of Translation in Prokaryotes
Initiation of translation
in Eukaryotes
It’s (much) more complex ..
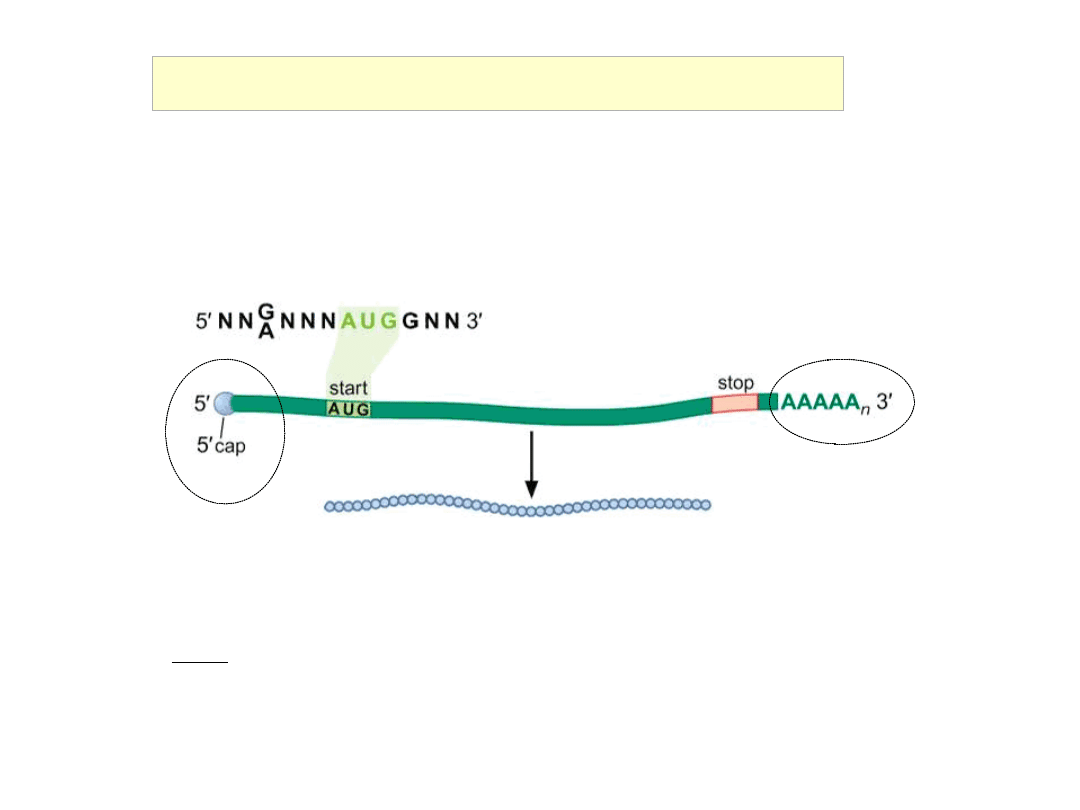
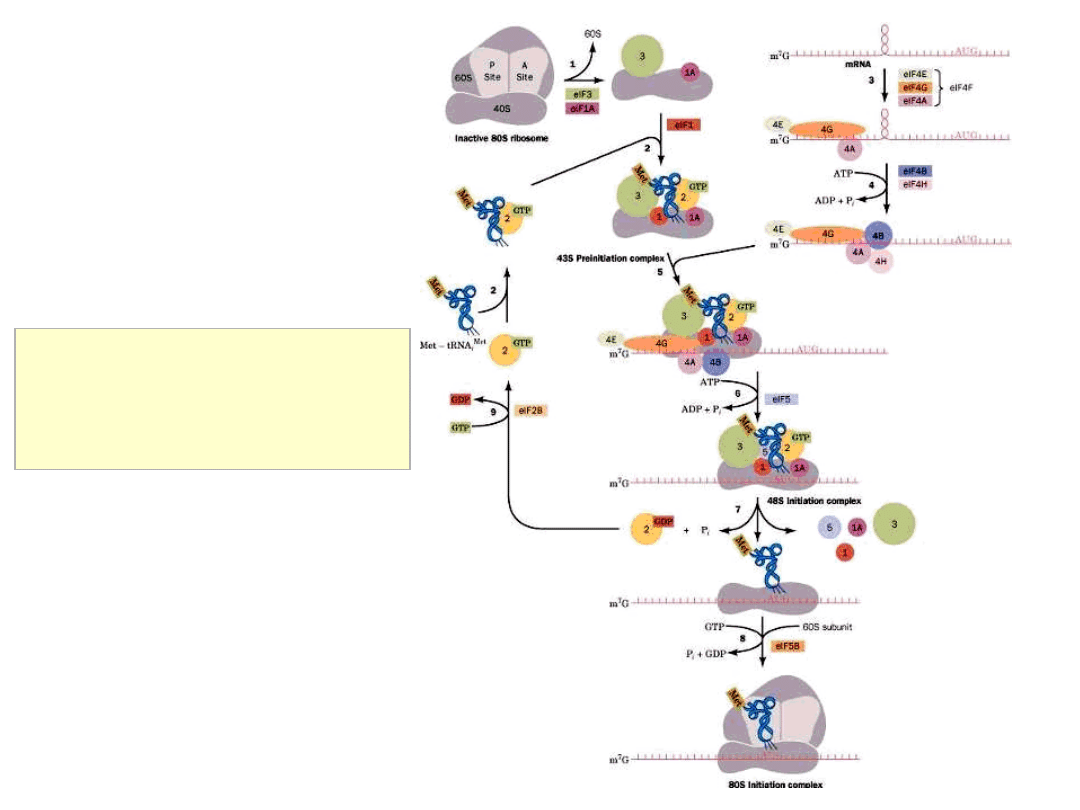
Initiation of Translation in Eukaryotes
major differences to prokaryotic mRNA……
• eukaryotic mRNAs possess a different 5’ ‘cap’ structure
• eukaryotic mRNAs are polyadenylated
Bases around the initiating AUG influence the efficiency of initiation:
RNNNAUGG (‘Kozak consensus’ sequence)
Eukaryotic initiation factor eIF4 scans along mRNA from cap to find
initiator AUG

(initiating)
AUG
open reading frame
A(n)
Stop codon
5’ ‘cap’
me7’
Gppp
43S
‘Scanning’ Model of Eukaryotic Initiation of Translation
(initiating)
AUG
open reading frame
A(n)
Stop codon
5’ ‘cap’
me7’
Gppp
‘scans’
(initiating)
AUG
A(n)
Stop codon
5’ ‘cap’
me7’
Gppp
(initiating)
AUG
A(n)
Stop codon
5’ ‘cap’
me7’
Gppp
60S
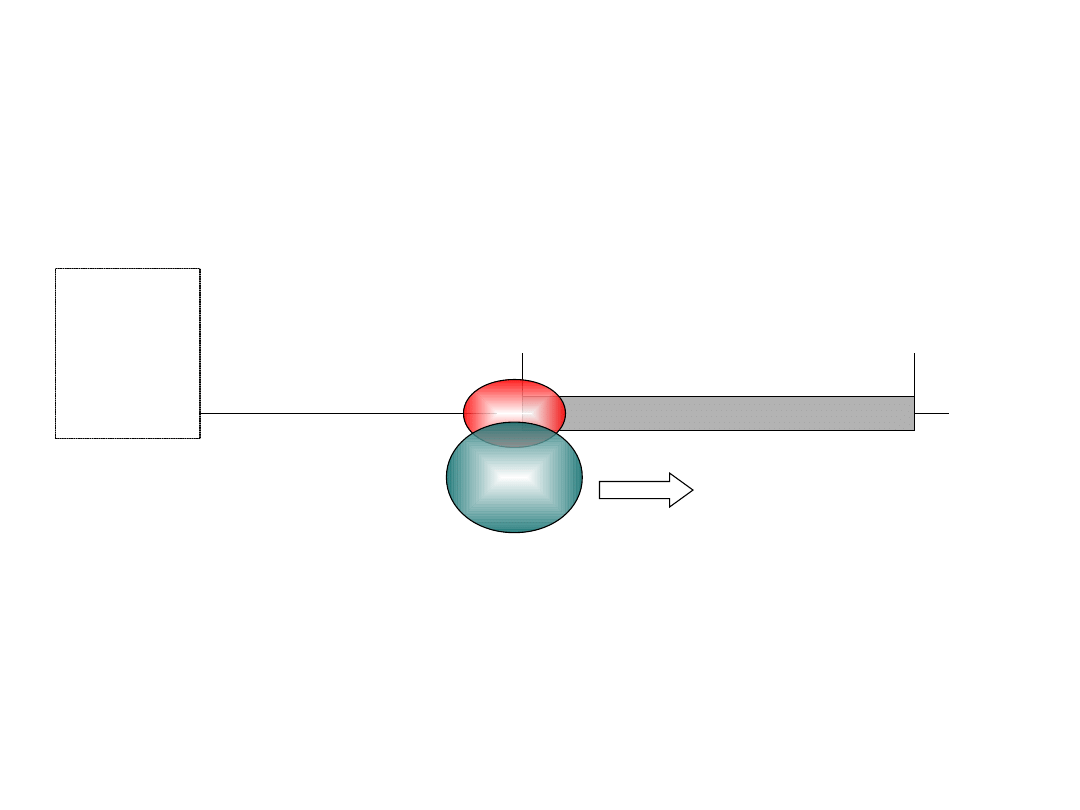
(initiating)
AUG
A(n)
Stop codon
5’ ‘cap’
me7’
Gppp
Elongation Phase
AAAAAAAAAAAAAAAA
m7Gppp
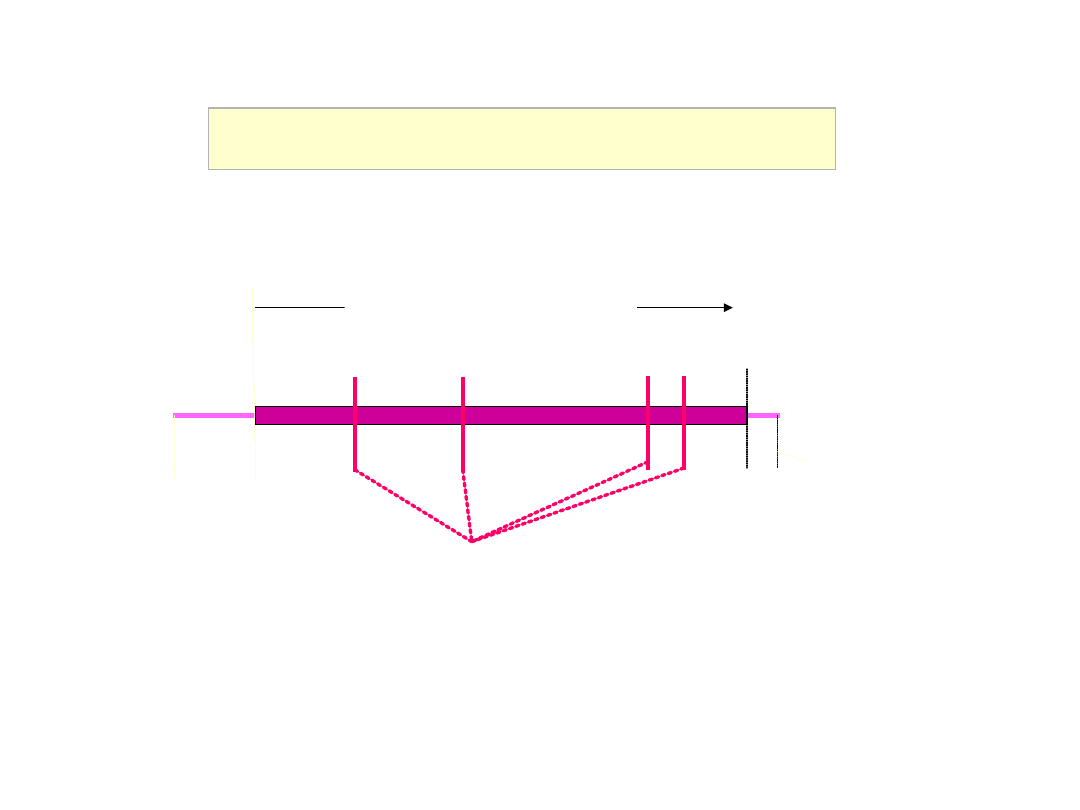
Messenger RNA structure
5’NCR
AUG
3’NCR
stop
Open reading frame
‘Cap’
Exon / exon splice boundaries
Poly(A) tail
m7Gppp
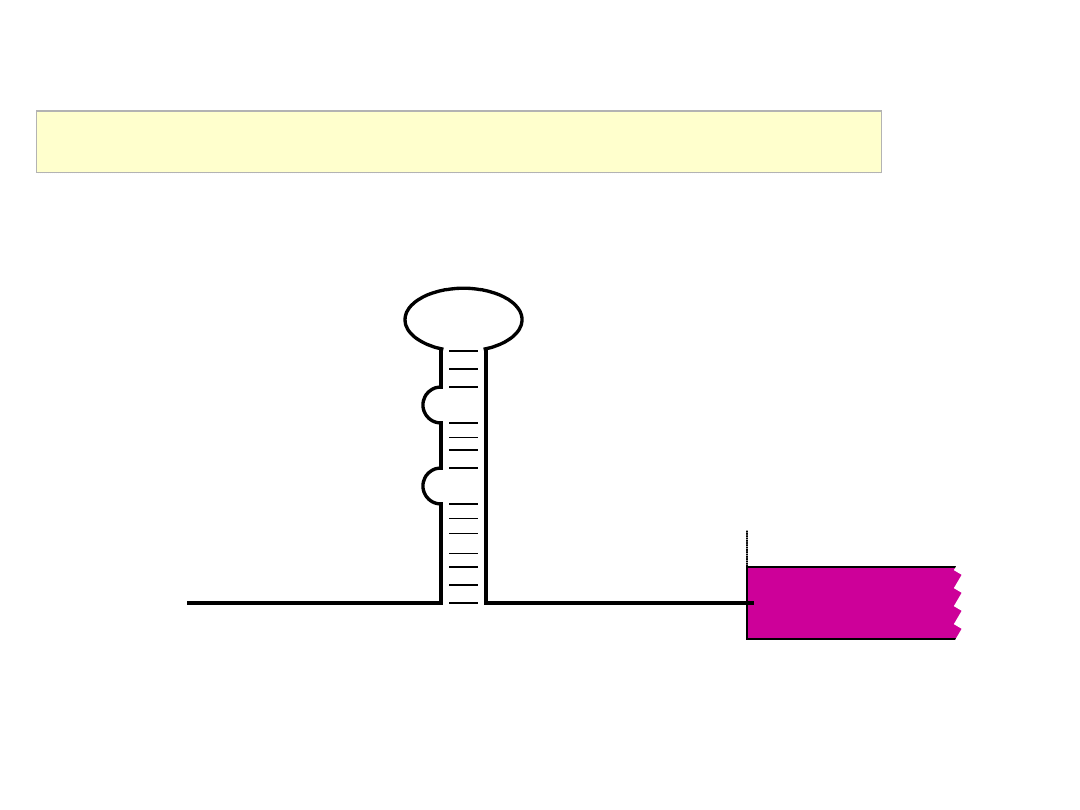
Messenger RNA structure – 5’ NCR
AUG
RNA stem-loop structures
AAAAAAAAAAAAA
orf
Messenger RNA structure – 3’ NCR
stop
A / U rich Elements (AREs)
- binding sites for stabilising
/ destabilising proteins
mRNA localisation elements
(usually located in the 3’NCR)
- binding sites for proteins which bind to the cytoskeleton
- binding sites for proteins (located at specific cellular sites)
which anchor the mRNA in that location
Initiation of Translation
60S
40S
40S
60S
60S
40S
+
eIF6
6
eIF3
3
Ribosome anti-association factor
Binds to 40S subunit (stabilises
Met-tRNAi) and prevents
association with 60S subunit
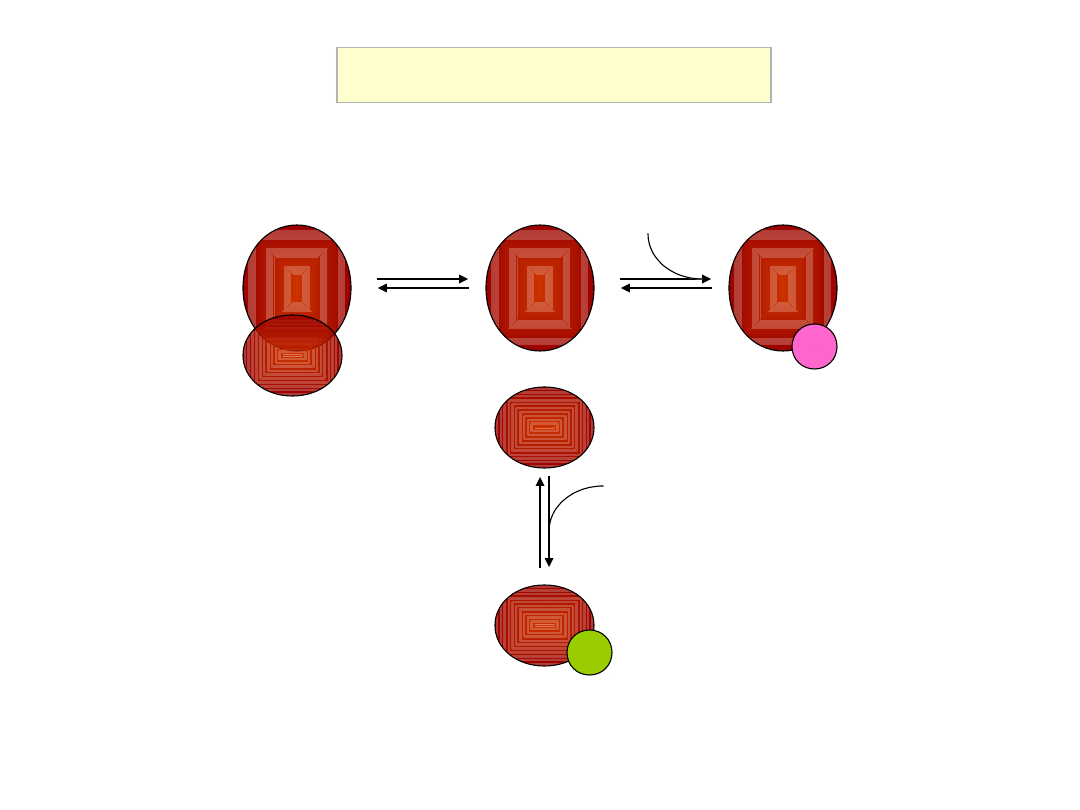
Ternary complex formation
eIF2
GDP
eIF2
GDP
eIF2B
eIF2
GTP
GTP
Met
eIF2
GTP
Met
eIF3
eIF5
ternary complex
eIF2
GTP
Met
multifactor complex (MFC)
eIF1
guanine nucleotide
exchange factor (GEF)
GDP-bound eIF2 cannot bind Met-tRNA
i
Met
GTPase activating protein (GAP)
initiator methionyl tRNA
eIF3
eIF5
eIF1
eIF3
eIF5
eIF2
GTP
Met
eIF1
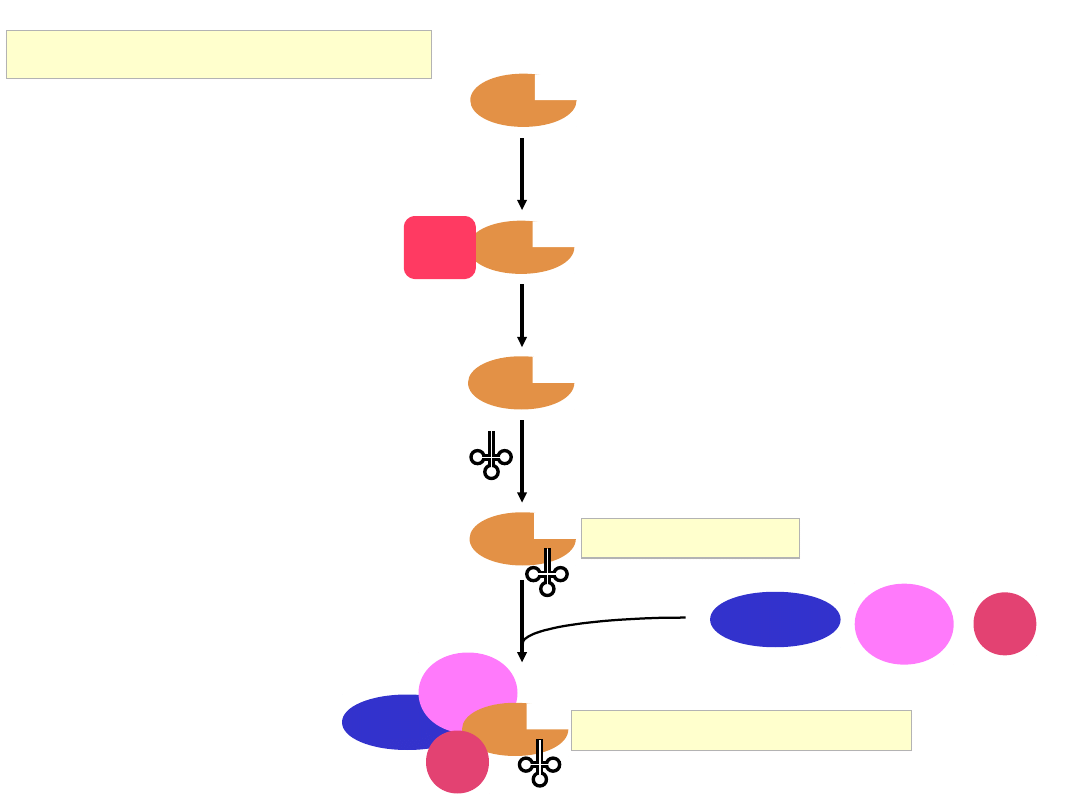
43S Complex Formation
eIF3
eIF5
eIF2
GTP
Met
eIF1
40S
eIF1A
eIF1A and eIF3 promote binding
of the multifactor complex to the
40S subunit
43S Complex
multifactor complex (MFC)
eIF1A
eIF4G
eIF4E
PABP
eIF4A
Binds poly(A) Tails
Binds 7meG ‘caps’
eIF4E : cap binding
eIF4A : bi-directional RNA helicase
MnkI : MAP kinase-interacting protein kinase-1 (phosphorylates eIF3)
PABP: poly(A) binding protein
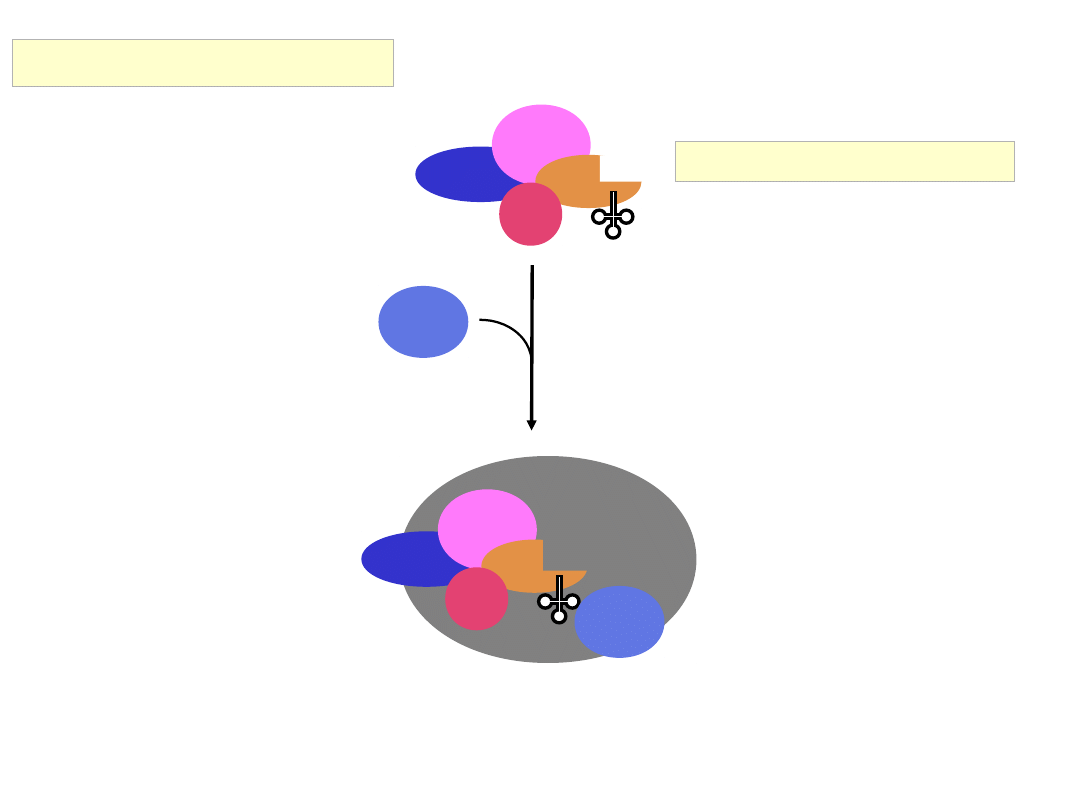
Assembly of the Cap Binding Complex:
Eukaryotic Initiation Factor 4G (eIF4G)
eIF3
eIF5
eIF2
GTP
Met
eIF1
40S
eIF1A
eIF4A
Binds
eIF3
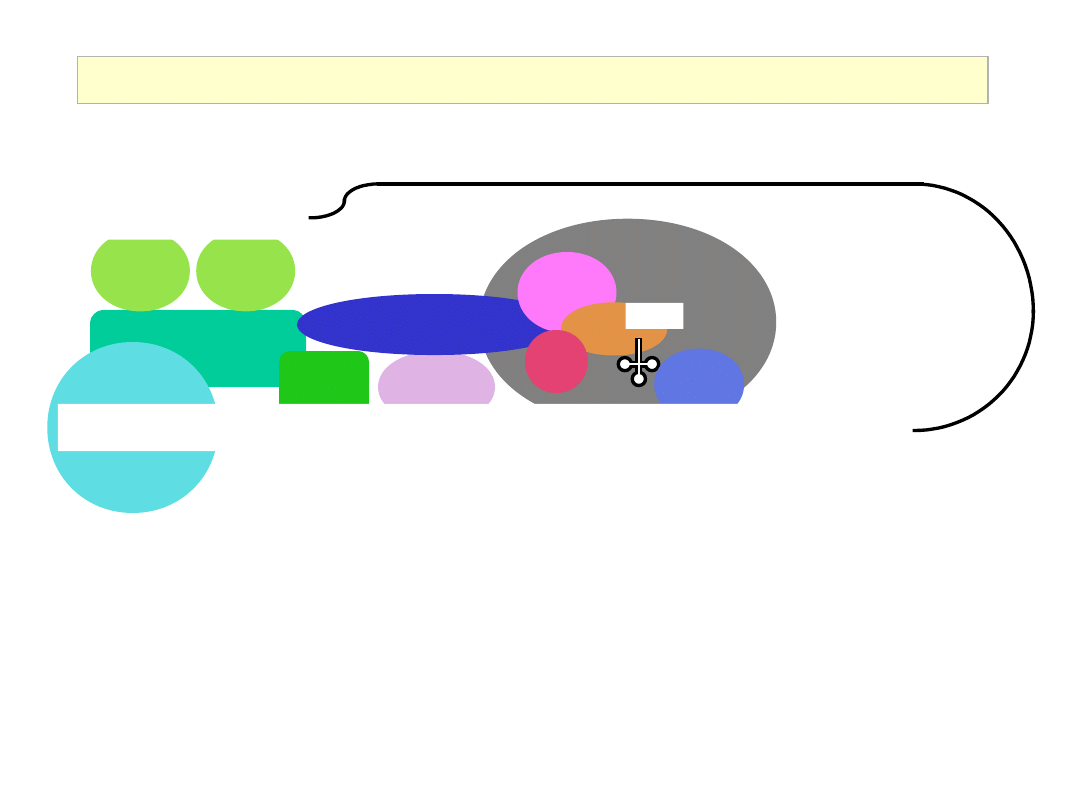
eIF4E
eIF4G
eIF4A
Recruitment of the 43S complex to the 5’ end of the mRNA
eIF4B
eIF3
eIF5
eIF2
GTP
Met
eIF1
40S
eIF1A
m
7
GpppGAUUCGAUACCAGGGAGCUUGGCACCAUGGC
• eIF3 : interacts with eIF4G to recruit the 43S complex
• eIF4B : RNA binding protein; stimulates (but not essential for) ribosome
binding to natural mRNA
PAB
P
PAB
P
AAAAAAAAA
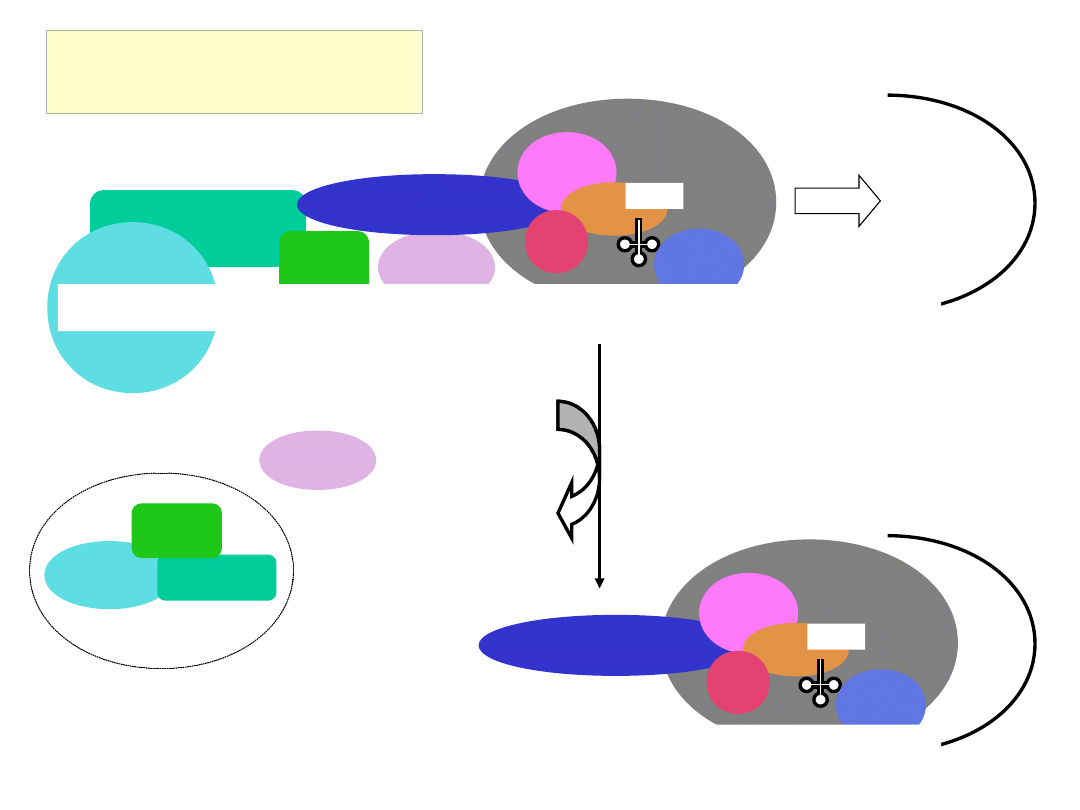
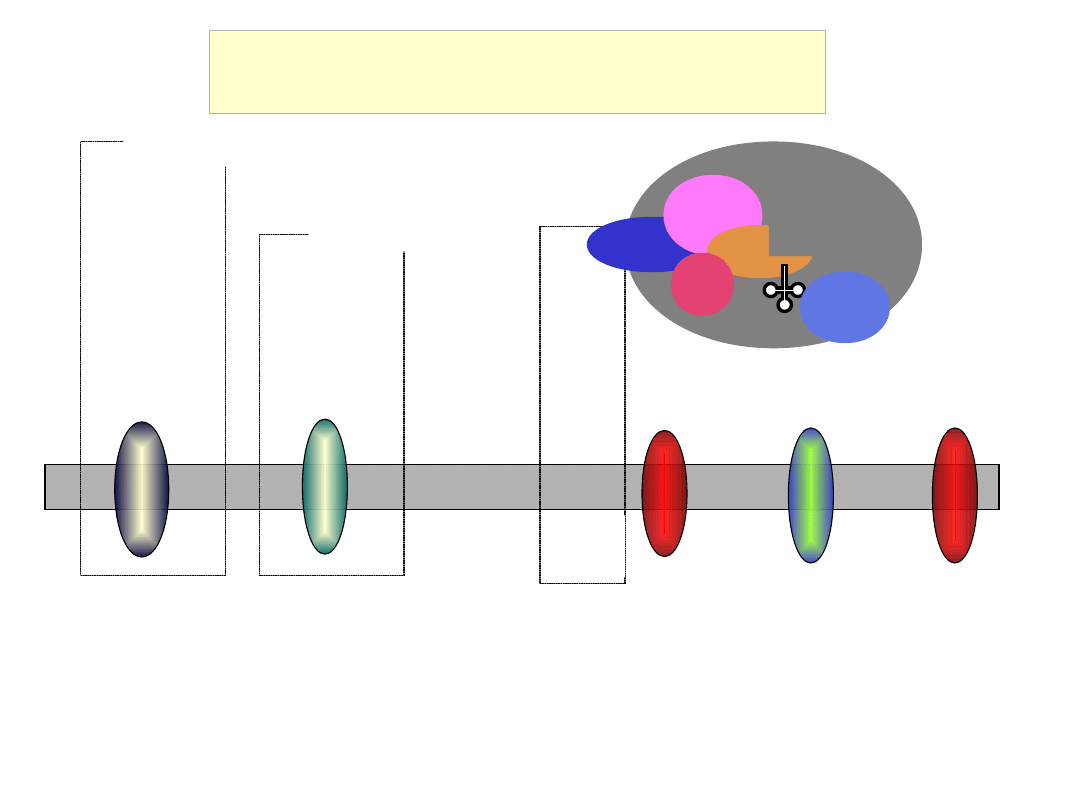
Scanning of the 5’ UTR
and AUG recognition
eIF4E
eIF4G
eIF4A eIF4B
eIF3
eIF5
eIF2
GTP
Met
eIF1
40S
eIF1A
m
7
GpppGAUUCGAUACCAGGGAGCUUGGC
ACC
AUG
G
C
eIF4E
eIF4G
eIF4A
eIF4B
eIF3
eIF5
eIF2
GTP
Met
eIF1
40S
eIF1A
m
7
GpppGAUUCGAUACCAGGGAGCUUGGC
ACC
AUG
G
C
ATP
ADP + P
i
cap-binding complex recycled
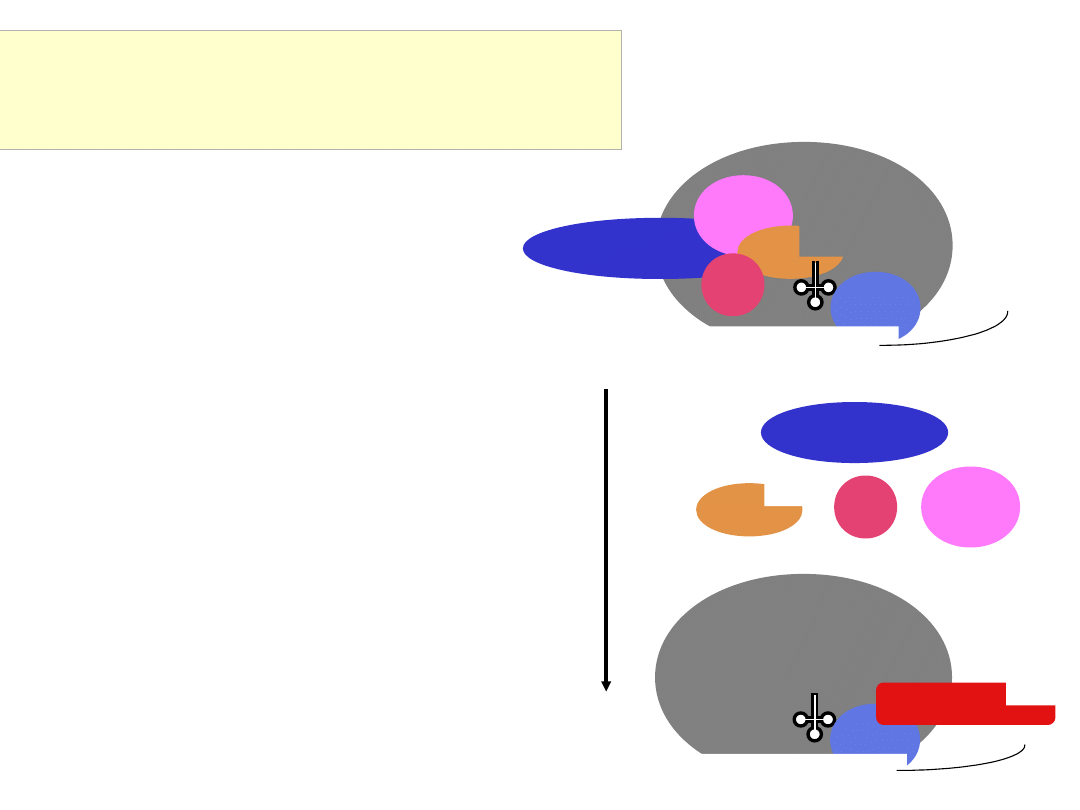
eIF3
eIF5
eIF2
GTP
Met
eIF1
40S
eIF1A
m
7
GpppGAUUCGAUACCAGGGAGCUUGGCACC
AUG
GC
Conformational change, GTP hydrolysis,
release of initiation factors,
and assembly of the eIF5B GTPase
eIF3
eIF5
eIF2
GDP
eIF1
Met
40S
eIF1A
m
7
GpppGAUUCGAUACCAGGGAGCUUGGCACC
AUG
GC
eIF5B
GTP
GTP hydrolysis by eIF2 requires eIF5 and possibly
a conformational change triggered by the
Met-tRNA
i
Met
interaction with the 40S subunit
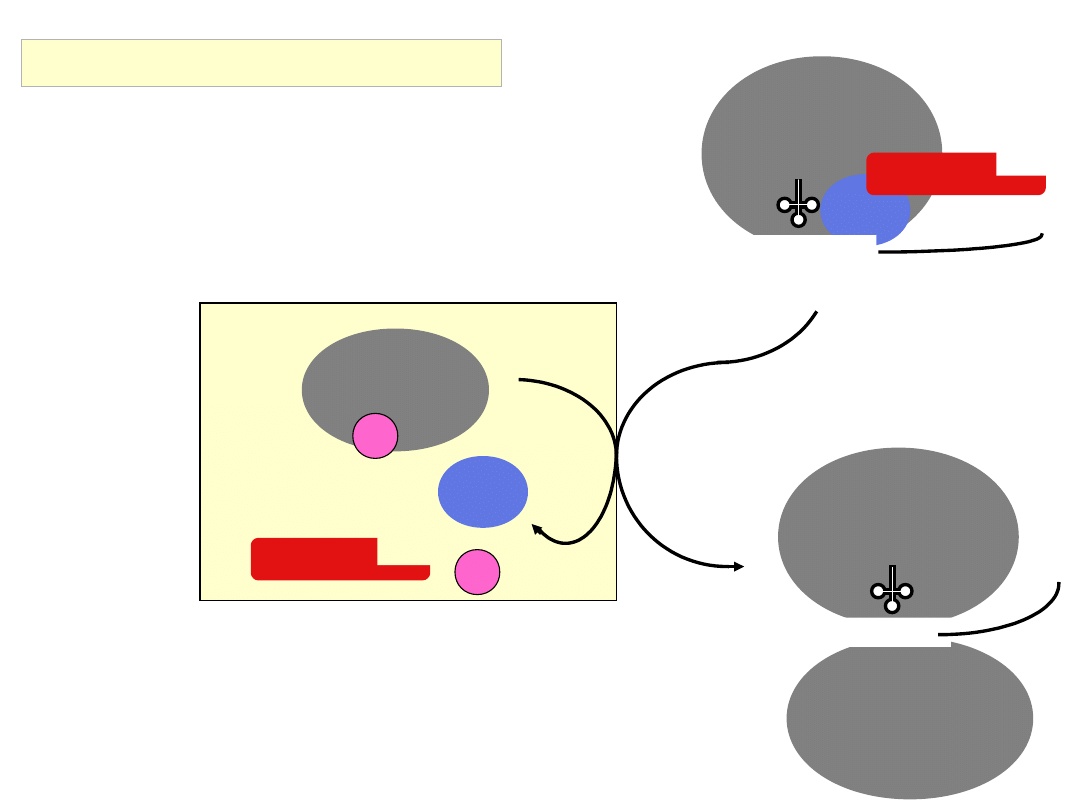
Met
40S
eIF1A
m
7
GpppGAUUCGAUACCAGGGAGCUUGGCACC
AUG
GC
eIF5B
GTP
Assembly of the 80S ribosome
40S
eIF1A
eIF5B
GDP
60S
GTP hydrolysis by eIF5B serves as a final
checkpoint for correct 80S assembly
the 80S ribosome is now poised to elongate
60
S
6
6
m
7
GpppGAUUCGAUACCAGGGAGCUUGGCACC
AUG
GC
Met
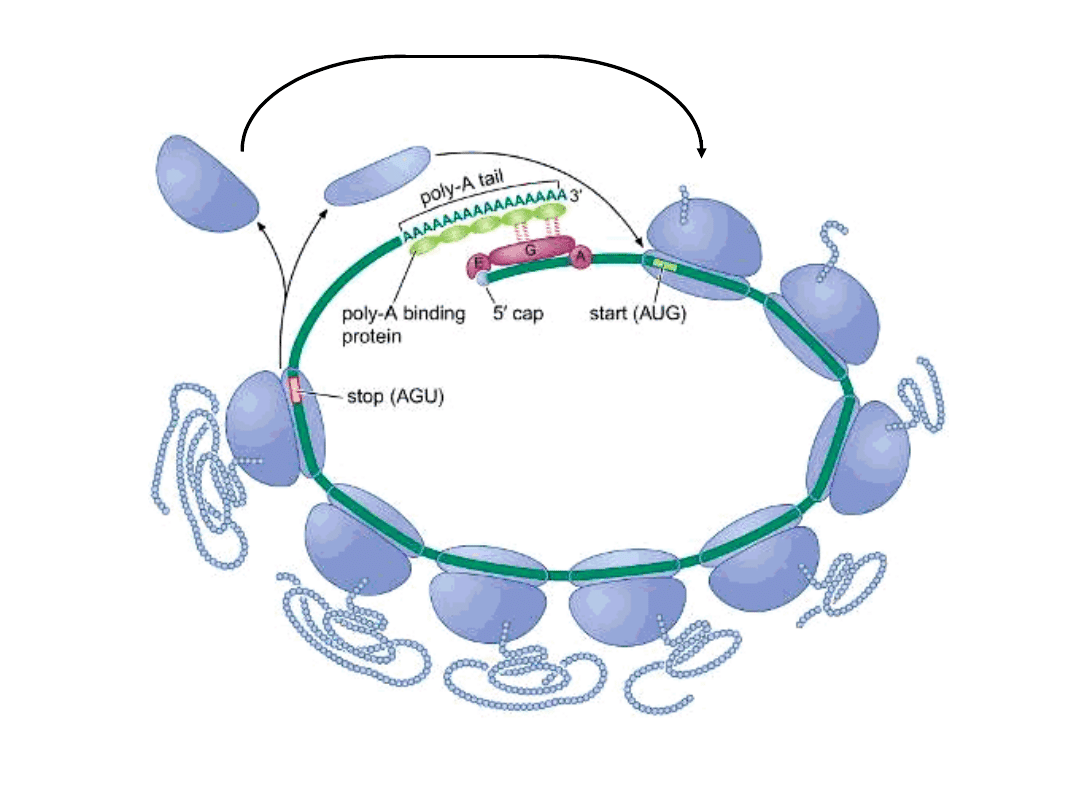

Initiation of Translation of Eukaryotic mRNAs
nothing similar to prokaryotic Shine-Dalgarno sequence
(ribosome binding
site; RBS)
5’ mRNA cap structure crucial
cap-binding protein complex recruits 43S complex
‘scanning’ of mRNA by 43S complex from 5’ cap structure to initiating
AUG
assembly of 80S ribosome ….. elongation
termination (proximal to site of initiation!!)
mRNAs translated as circular complexes
(essentially) no mechanism for internal initiation – eukaryotic mRNAs
are monocistronic

Factor Mol mass (kDa) and Function (based on biochemical studies in mammals)
subunit composition
eIF-1 14 Omission restricts binding of 40S subunit to 5’ end (entry
site)
on mRNA
eIF-1A 17 Catalytically promotes Met-tRNAi binding to 40S;
required for
strong binding of 40S subunit to mRNA
eIF-2
36 (), 38 (), 52 () GTP binding protein; escorts Met-tRNAi onto 40S
ribosomal
subunit
eIF-2B
81, 71, 58, 43, 34 Guanine nucleotide exchange factor (GEF): promotes
exchange
of GDP for GTP on eIF-2
eIF-3
110, 67, 42, 40, 36, 35 Binds to 40S subunit, stabilizing Met-tRNAi and
preventing
association with 60S subunit
eIF-4E
25 Binds directly to m7G cap
eIF-4G 220 Augments binding of eIF-4E to m7G cap; required for the
initial
round but not for sustained translation
eIF-4A 46 RNA-dependent ATPase; essential for binding of
ribosomes to
natural mRNA
eIF-4B 69 RNA binding protein; stimulates (but not essential for)
ribosome
binding to natural mRNA
eIF-5
45 Mediates hydrolysis of GTP associated with eIF-2 on 40S
subunit
(eIF-6 25kDa Ribosome anti-association factor)

The Elongation Phase of Translation
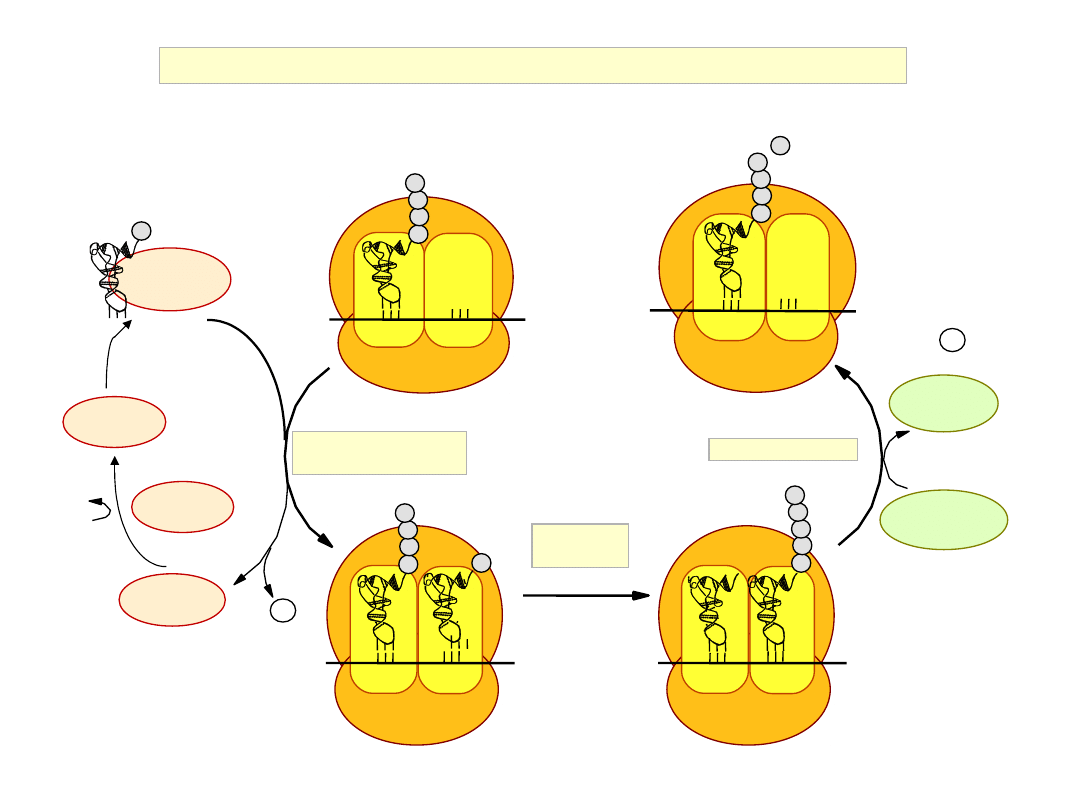
Elongation Cycle of Eukaryotic Protein Synthesis
A
P
5'
A
P
5'
An
aa
aa
aa
aa
aa
aa
aa
aa
aa
aa
peptidyl
transfer
aminoacyl-tRNA
binding
EF1A GTP
P
EF1A GDP
EF1A GTP
EF1B
GDP
GTP
An
A
P
5'
An
A
P
5'
aa
aa
aa
aa
aa
aa
aa
aa
aa
aa
EF2 GDP
EF2 GTP
P
+
Translocation
An
Elongation factor 2
- a molecular motor
tRNA

The Termination Phase of Translation
Human
eRF1
tRNA
CCA acceptor stem
anti-codon loop
anticodon-like site
-TASNIKS-
• terminates translation
• recognises all three stop codons
• activates a water molecule to hydrolyse tRNA-peptide ester
linkage
eRF1
- GGQ -
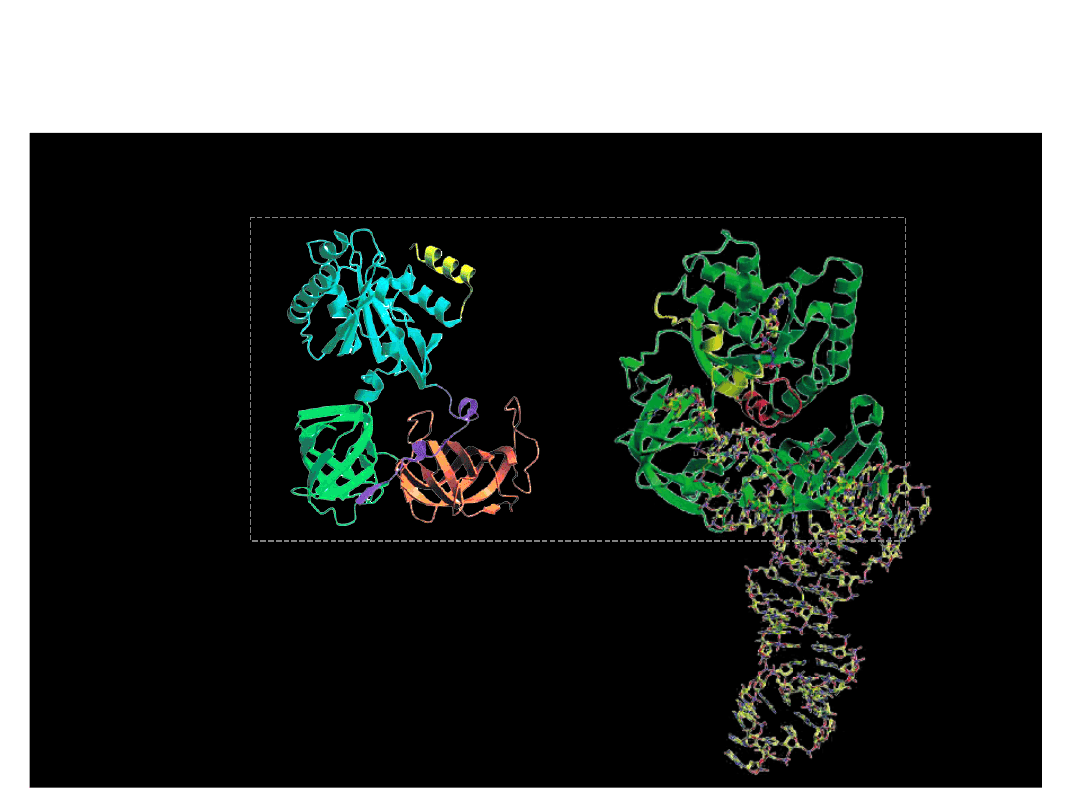
tRNA
EF-tu (eEF1A)
eRF3
GTPase : enhances termination efficiency
stimulates eRF1 activity in a GTP-dependent
manner
eRF3: a molecular mimic of elongation factor 1A
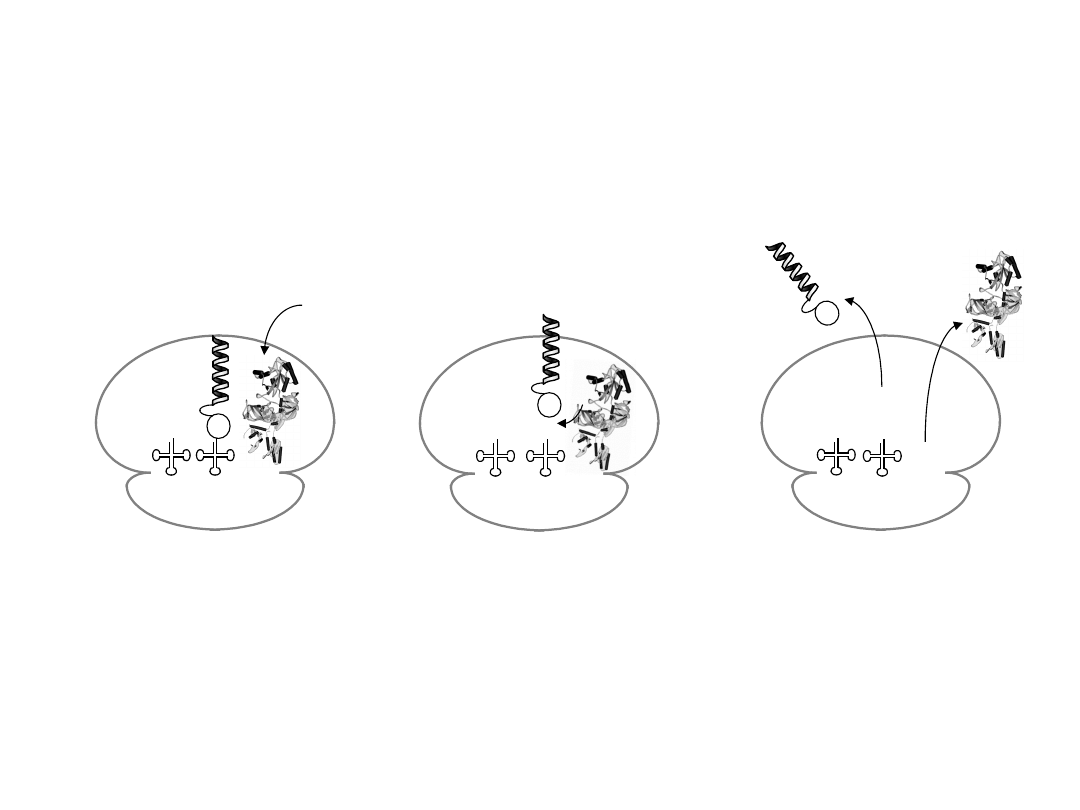
E P A
G
E P A
OH
OH
E P A
G
OH
OH
G
OH
eRF1
H
2
0
eRF1 binds into A site
(interacts with stop codon)
eRF1 activates a water
molecule to hydrolyse the
peptidyl-tRNA ester linkage
eRF3 binds and accelerates
the dissociation of eRF1
- nascent protein released

Document Outline
- Slide 1
- Slide 2
- Slide 3
- Slide 4
- Slide 5
- Slide 6
- Slide 7
- Slide 8
- Slide 9
- Slide 10
- Slide 11
- Slide 12
- Slide 13
- Slide 14
- Slide 15
- Slide 16
- Slide 17
- Slide 18
- Slide 19
- Slide 20
- Slide 21
- Slide 22
- Slide 23
- Slide 24
- Slide 25
- Slide 26
- Slide 27
- Slide 28
- Slide 29
- Slide 30
- Slide 31
- Slide 32
- Slide 33
- Slide 34
- Slide 35
- Slide 36
- Slide 37
- Slide 38
- Slide 39
- Slide 40
Wyszukiwarka
Podobne podstrony:
2010 lecture 3 transl groups
2010 Lecture 6, 7 transl groups
2010 lecture 4 transl groups
2010 lecture 5 transl groups
2010 lecture 2 transl groups
2010 lecture 10 transl groups medieval theatre
IR Lecture1
uml LECTURE
lecture3 complexity introduction
33 Przebieg i regulacja procesu translacji
196 Capital structure Intro lecture 1id 18514 ppt
Lecture VIII Morphology
BM7 Translacja
benzen lecture
lecture 1
Lecture10 Medieval women and private sphere
8 Intro to lg socio1 LECTURE2014
lecture 3
więcej podobnych podstron