
Biochemia 2008/09
TRANSLACJA

Messenger RNA (mRNA)
m
7
Gppp
Cap
5’
5’ untranslated region
AUG
initiation
codon
translated (coding) region
(AAAA)
n
poly(A)
tail
3’ untranslated region
UGA
termination
codon
3’
AAUAAA
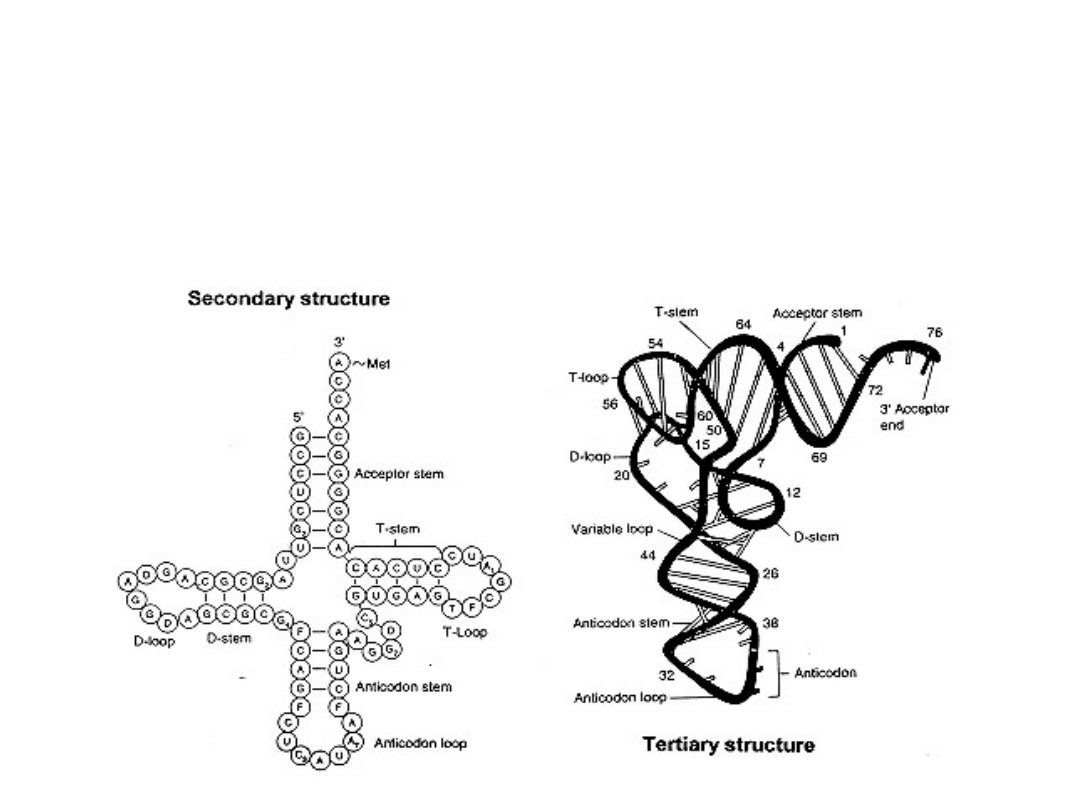
Transfer RNA
• tRNA is the “adaptor” molecule in protein synthesis
• acceptor stem
• CCA-3’ terminus to which amino acid is coupled
• carries amino acid on terminal adenosine
•anticodon stem and anticodon loop

Amino acid activation
and aminoacyl tRNA synthetases
• aminoacyl tRNA synthetases are the enzymes that “charge” the tRNAs
• 20 amino acids
• one aminoacyl tRNA synthetase for each amino acid
• can be several different “isoacceptor” tRNAs for each amino acid
• all isoacceptor tRNAs for an amino acid use the same synthetase
each aminoacyl tRNA synthetase binds
• amino acid
• ATP
• isoacceptor tRNAs
H
2
N-C-C-OH
H
R
-
-
O
=
ATP
H
2
N-C-C-O-P-O-ribose-adenine
H
R
-
-
O
=
amino acid
adenylated (activated)
amino acid
PPi
uncharged tRNA
H
2
N-C-C-O
H
R
-
-
O
=
aminoacyl
(charged)
tRNA
AMP
3’
Amino acid activation
and
tRNA charging

The genetic code
• consists of 64 triplet codons (A, G, C, U) 4
3
= 64
• all codons are used in protein synthesis
• 20 amino acids
• 3 termination (stop) codons: UAA, UAG, UGA
• AUG (methionine) is the start codon (also used internally)
• multiple codons for a single amino acid =
degeneracy
• 5 amino acids are specified by the first two nucleotides only
• 3 additional amino acids (Arg, Leu, and Ser) are specified by
six different codons
The Genetic
Code
UUU
UUC
UUA
UUG
CUU
CUC
CUA
CUG
AUU
AUC
AUA
AUG
GUU
GUC
GUA
GUG
UCU
UCC
UCA
UCG
CCU
CCC
CCA
CCG
ACU
ACC
ACA
ACG
GCU
GCC
GCA
GCG
UAU
UAC
UAA
UAG
CAU
CAC
CAA
CAG
AAU
AAC
AAA
AAG
GAU
GAC
GAA
GAG
UGU
UGC
UGA
UGG
CGU
CGC
CGA
CGG
AGU
AGC
AGA
AGG
GGU
GGC
GGA
GGG
Phe
Leu
Leu
Val
Ile
Met
Ser
Pro
Thr
Ala
Tyr
Stop
His
Gln
Asn
Lys
Asp
Glu
Cys
Arg
Ser
Arg
Gly
Stop
Trp
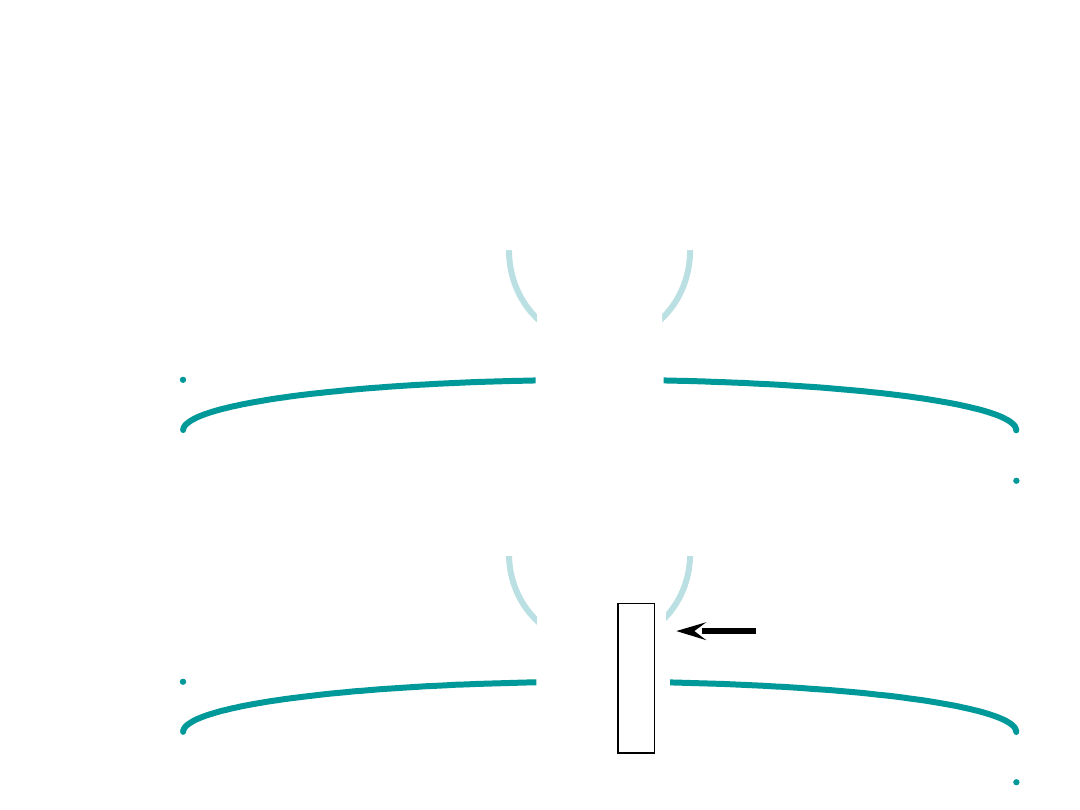
Codon-anticodon interactions
• codon-anticodon base-pairing is antiparallel
• the third position in the codon is frequently degenerate
• one tRNA can interact with more than one codon (therefore 50 tRNAs)
• wobble rules
• C with G or I (inosine)
• A with U or I
• G with C or U
• U with A, G, or I
• I with C, U, or A
5’
3’
A U G
U A C
3’
5’ tRNA
met
mRNA
5’
3’
C U A
G
G A U
3’
5’ tRNA
leu
mRNA
wobble base
• one tRNA
leu
can read two
of the leucine codons
Wobble Interactions

Cechy kodu:
Zdegerowany, niedwuznaczny,
nie nakładający się, uniwersalny,
bezprzestankowy
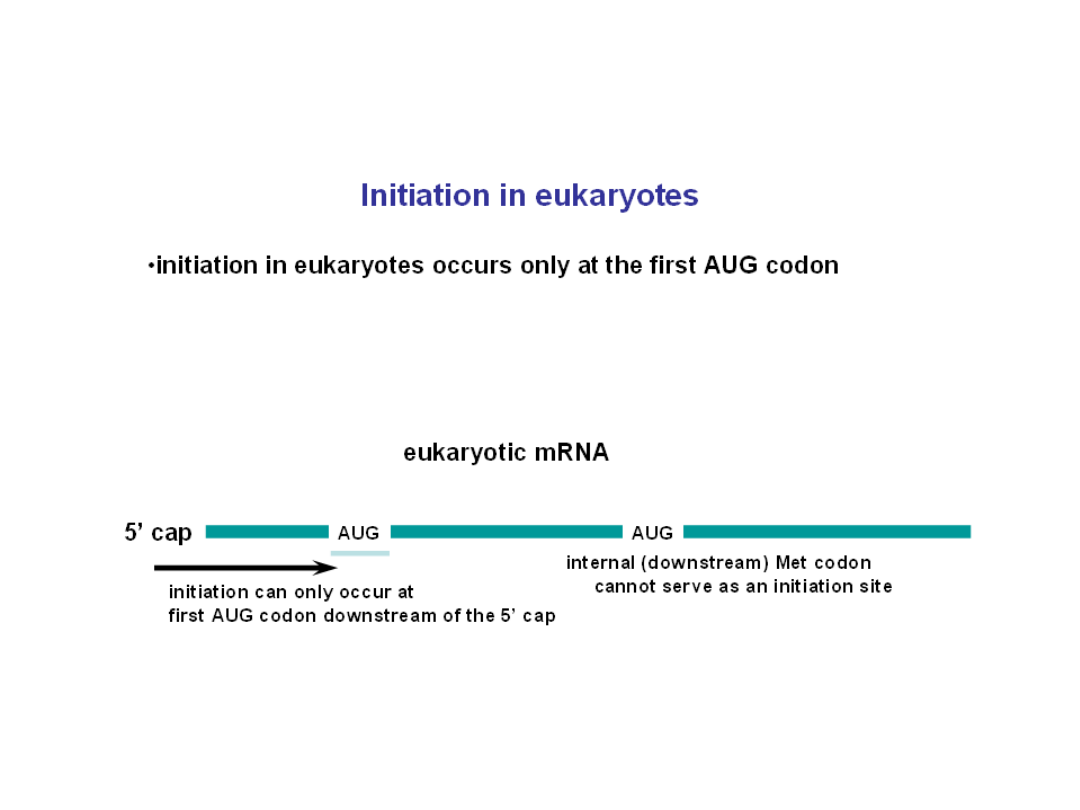

Reading frame
• reading frame is determined by the AUG initiation codon
• every subsequent triplet is read as a codon until reaching a stop codon
...AGAGCGGA.
AUG.GCA.GAG.UGG.CUA.AGC.AUG.UCG.
UGA.UCGAAUAAA...
MET.ALA.GLU.TRP.LEU.SER.MET.SER
• a frameshift mutation
...AGAGCGGA.AUG.GCA.GA .UGG.CUA.AGC.AUG.UCG.UGA.UCGAAUAAA...
• the new reading frame results in the wrong amino acid sequence and
the formation of a truncated protein
...AGAGCGGA.
AUG.GCA.GAU.GGC.
UAA.GCAUGUCGUGAUCGAAUAAA...
MET.ALA.
ASP.GLY
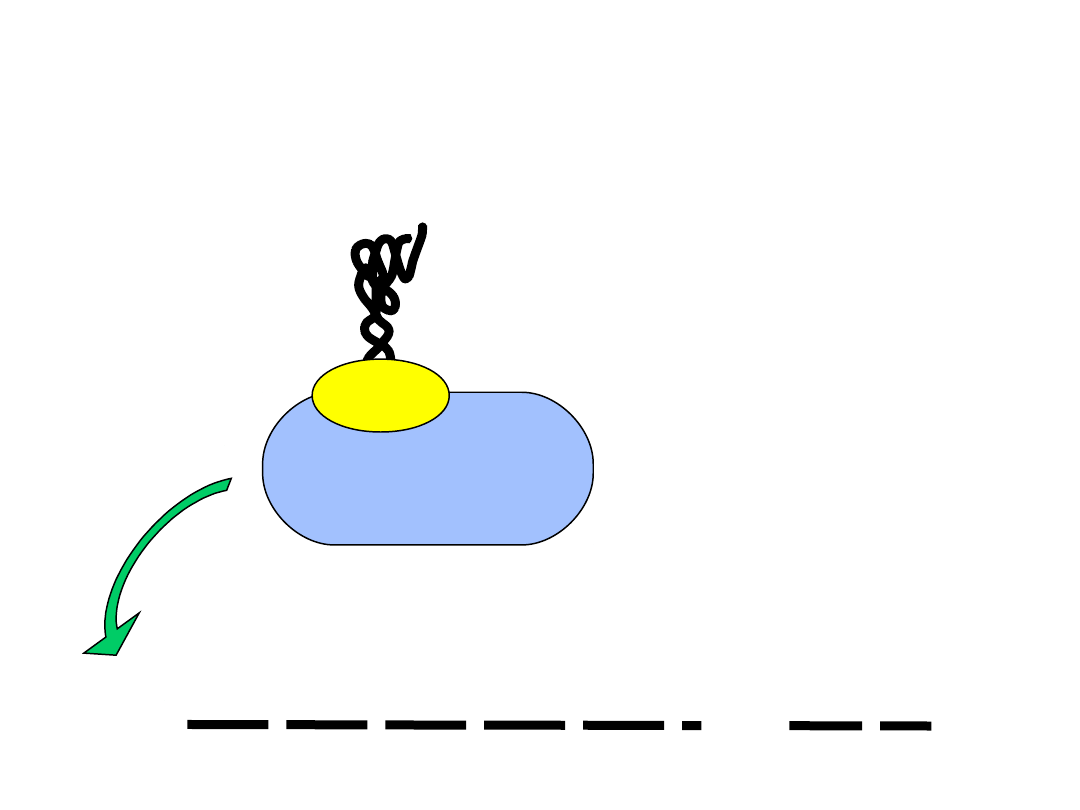
mRNA
5’ cap
40S subunit
M
eIF2
AUG
Initiator tRNA bound to the
small ribosomal subunit with the
eukaryotic initiation factor-2 (eIF2)
Initiation of protein synthesis: mRNA binding
The small subunit finds the 5’ cap and
scans down the mRNA to the first AUG codon
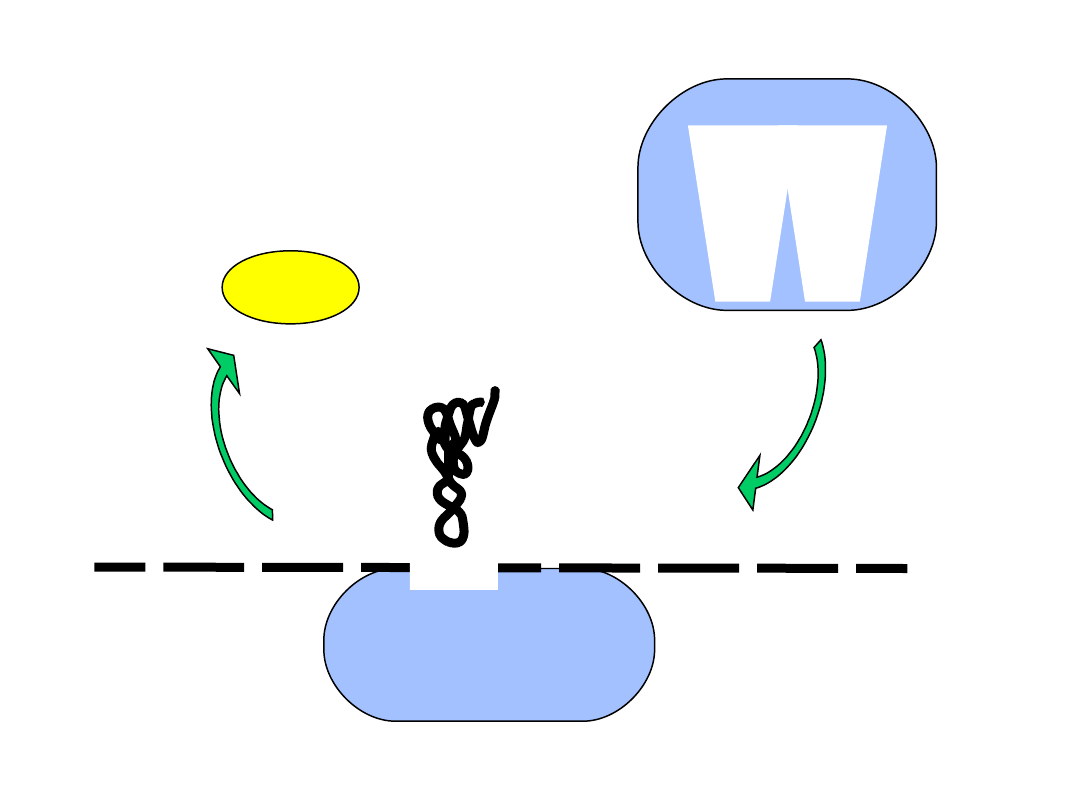
mRNA
5’
40S subunit
M
eIF2
AUG
• the initiation codon is recognized
• eIF2 dissociates from the complex
• the large ribosomal subunit binds
60S subunit
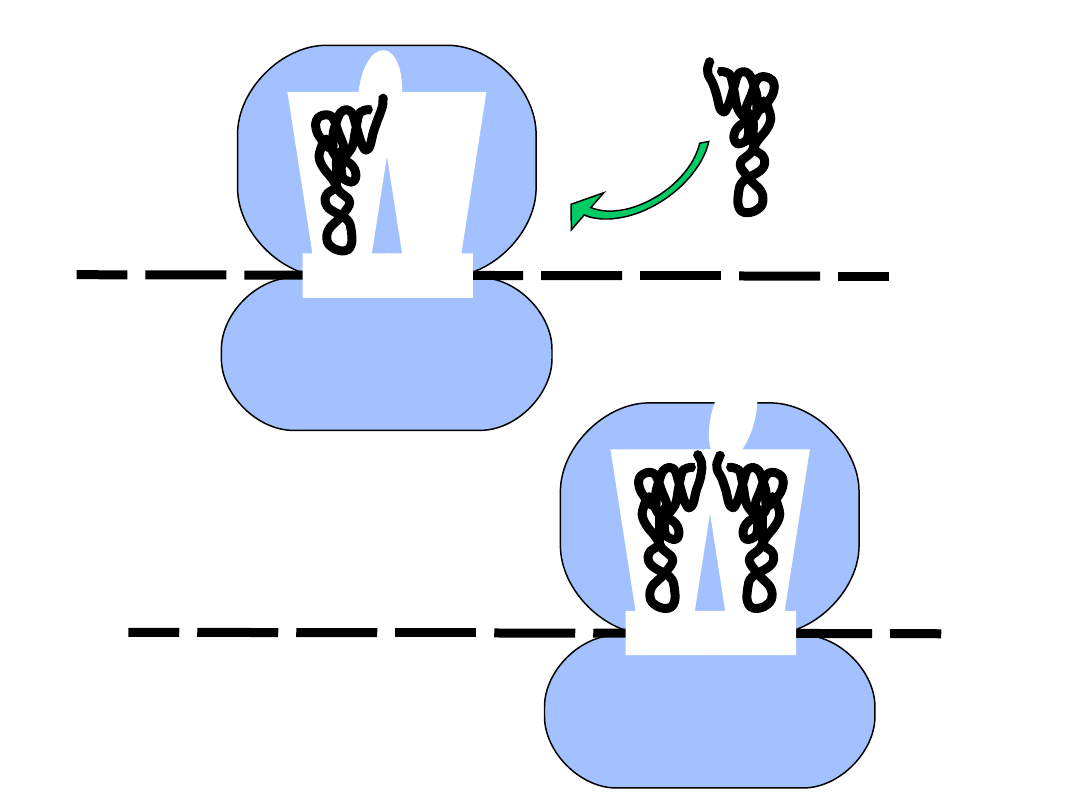
mRNA
5’
M
AUG
• aminoacyl tRNA binds the A-site
• first peptide bond is formed
GCC
A
mRNA
5’
M
AUG
GCC
A
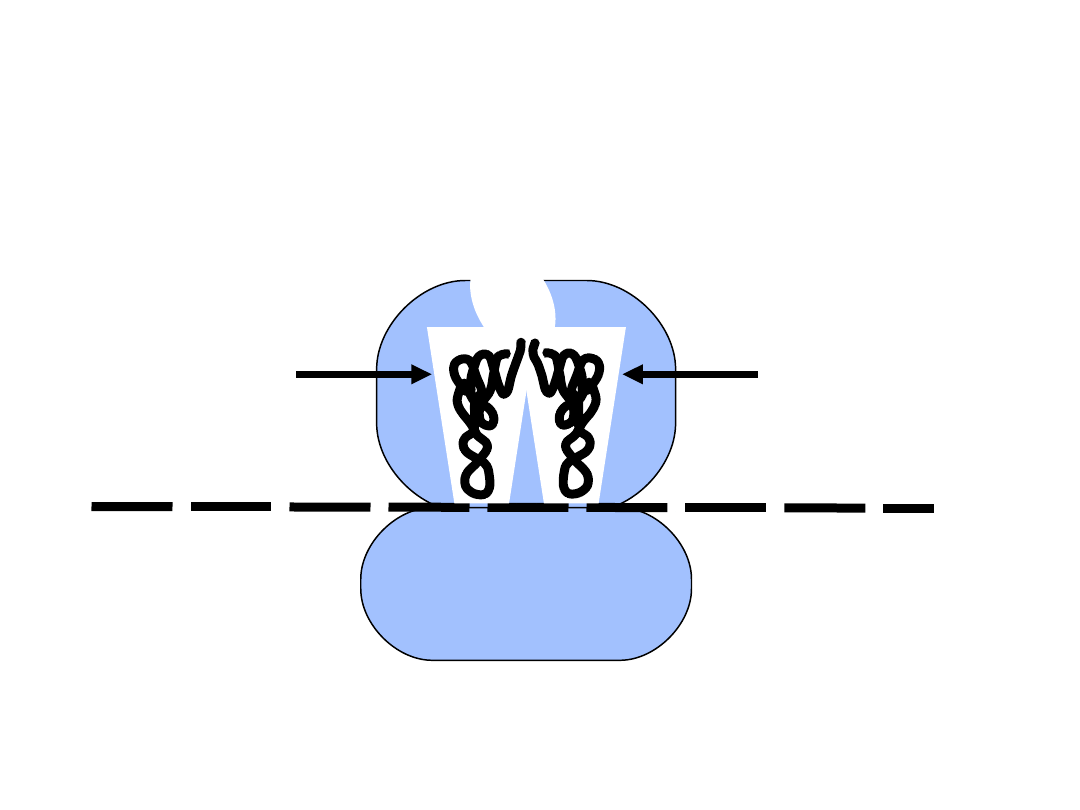
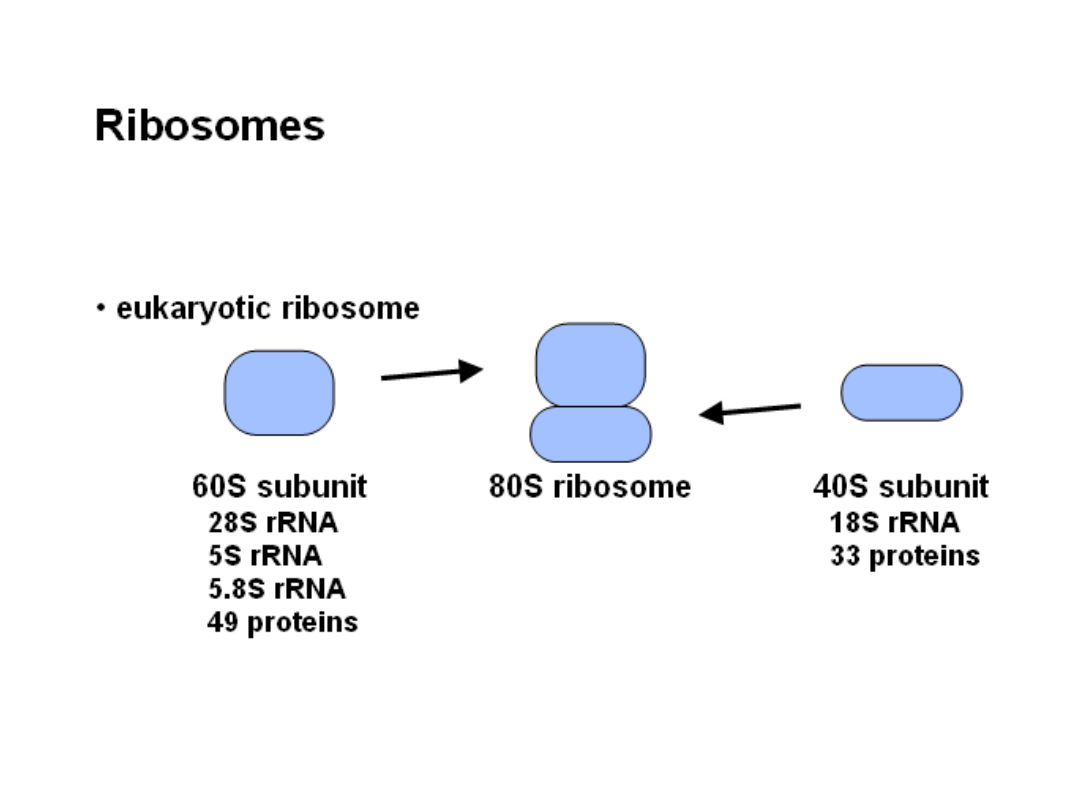
Ribosome structure
A
P PP
P
P
P
P
P
P-site
peptidyl tRNA site
A-site
aminoacyl tRNA site
mRNA
5’
Small subunit
Large subunit
Ribosome with bound tRNAs and mRNA
C
NH
2
CH
3
-S-CH
2
-CH
2
-CH
O=C
Peptide bond formation
• peptide bond formation is
catalyzed by
peptidyl transferase
•
peptidyl transferase is contained within
a sequence of 28S rRNA in the
large ribosomal subunit
;
• the energy for peptide bond formation
comes from the ATP used in tRNA charging
• peptide bond formation results in a shift
of the nascent peptide from the P-site
to the A-site
NH
2
CH
3
-S-CH
2
-CH
2
-CH
O=C
O
tRNA
NH
2
CH
3
-CH
O=C
O
tRNA
N
P-site
A-site
OH
tRNA
NH
CH
3
-CH
O=C
O
tRNA
P
UCA
P
P
P
P
P
UCA GCA GGG UAG
A
P
P
P
P
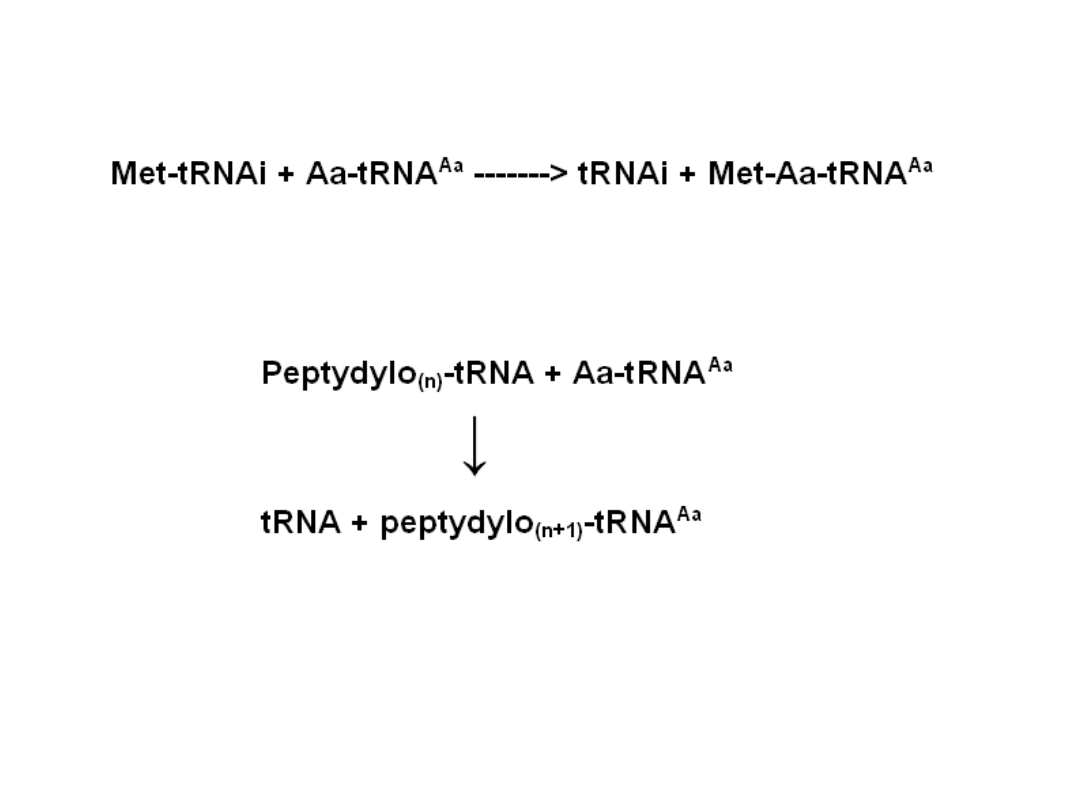
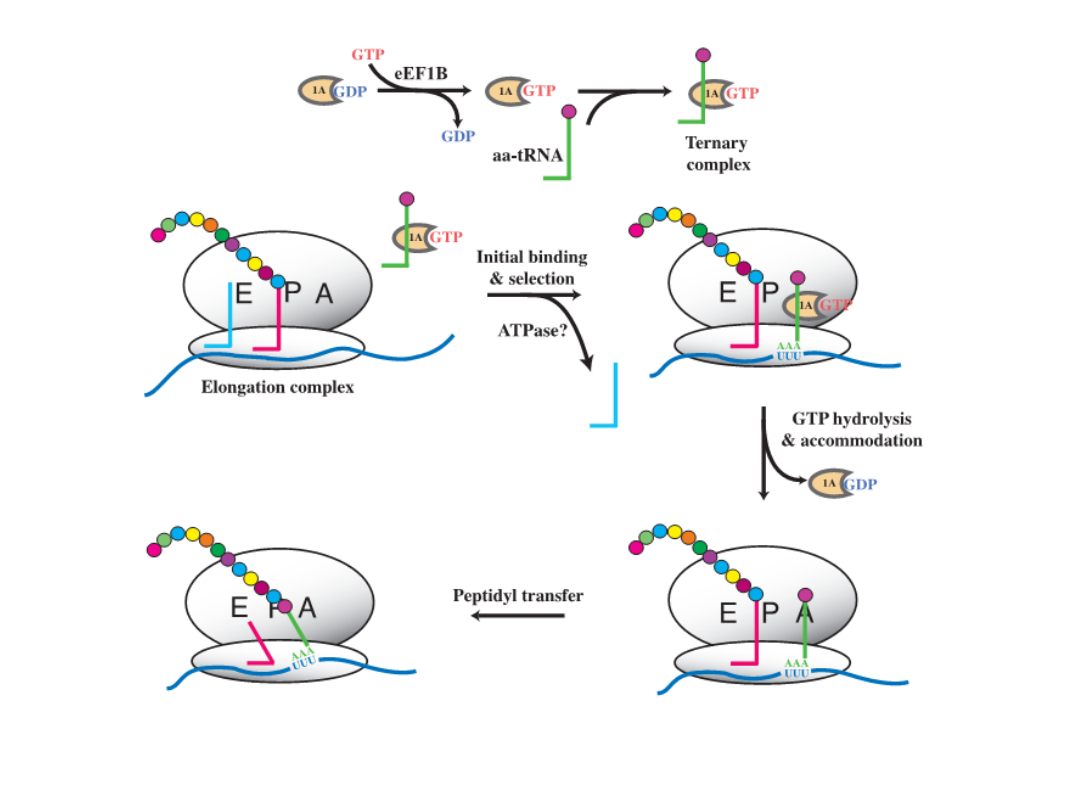
Elongation
GCA GGG UAG
• following peptide bond formation
the uncharged tRNA dissociates
from the P-site
• the ribosome shifts one codon along
the mRNA, moving peptidyl tRNA
from the A-site to the P-site; this
translocation requires the
elongation factor EF2
• the next aminoacyl tRNA then
binds within the A-site; this tRNA
binding requires the elongation
factor EF1
• energy for elongation is provided by
the hydrolysis of two GTPs:
• one for translocation
• one for aminoacyl tRNA binding
EF1
EF2
P
UCA GCA GGG UAG
P
P
P
P
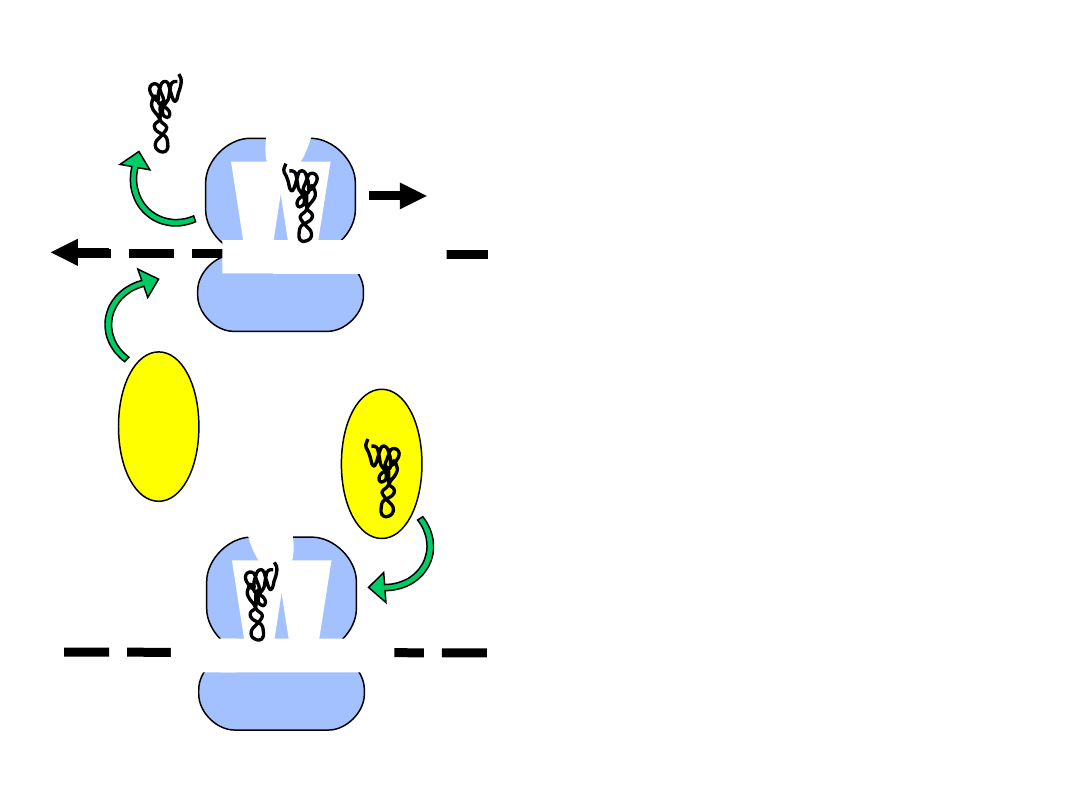
Termination
• when translation reaches the stop
codon, a release factor (RF) binds
within the A-site, recognizing the
stop codon
• release factor catalyzes the hydrolysis
of the completed polypeptide from
the peptidyl tRNA, and the entire
complex dissociates
RF
P
UCA GCA GGG UAG
P
P
P
P
P
P
P
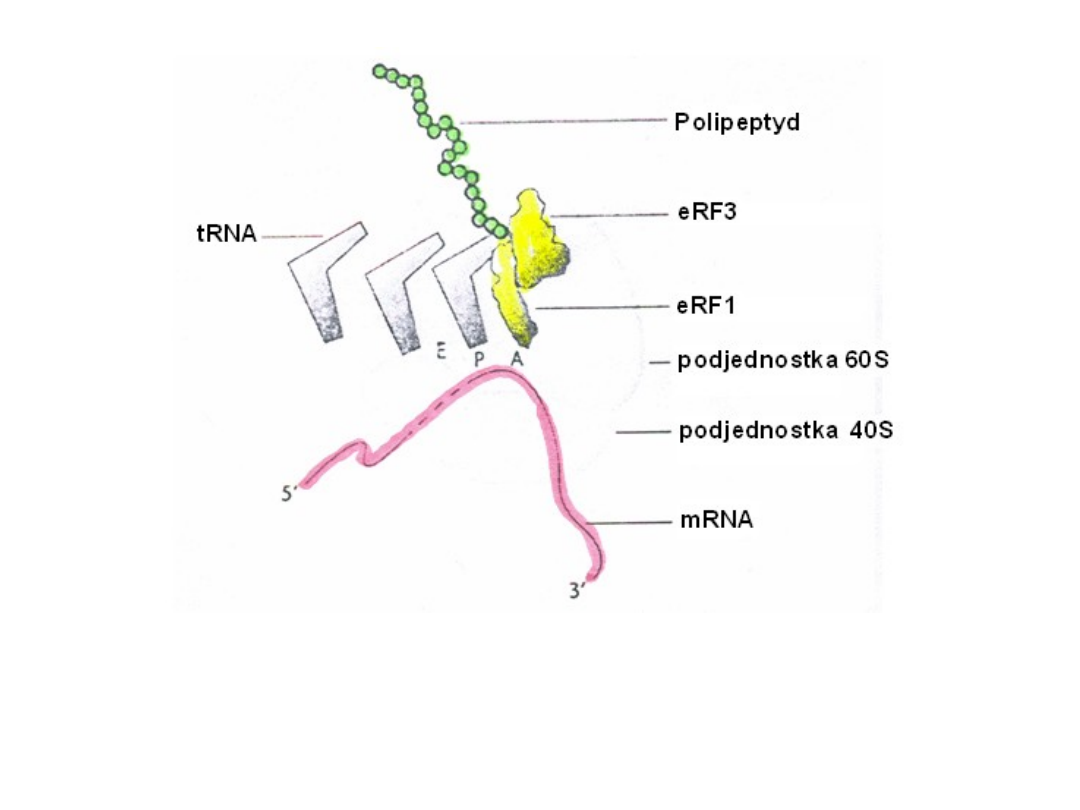
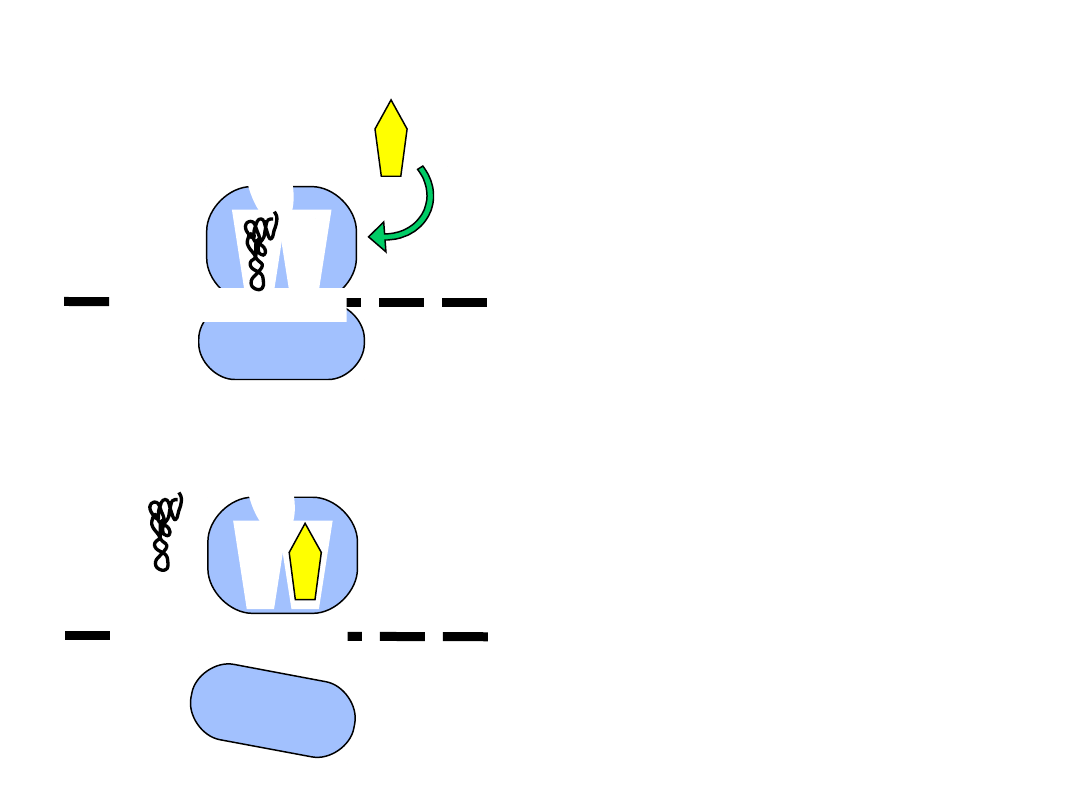
Składniki reakcji terminacji translacji
w komórkach eukariotycznych
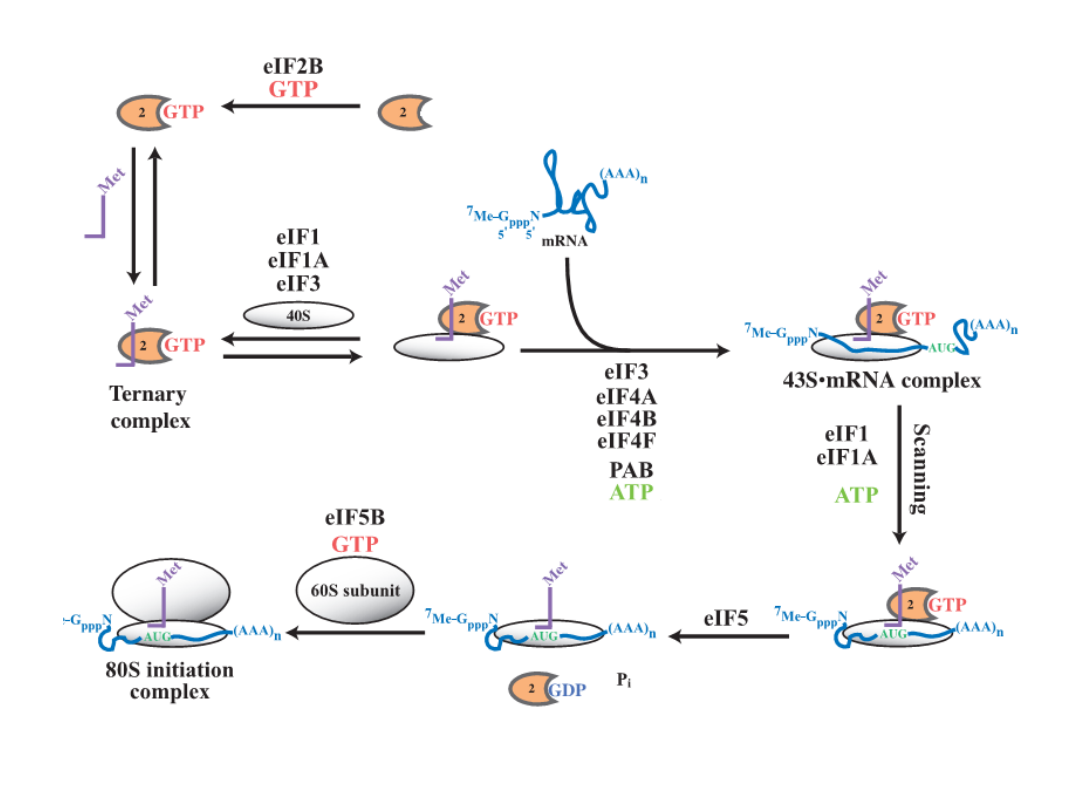
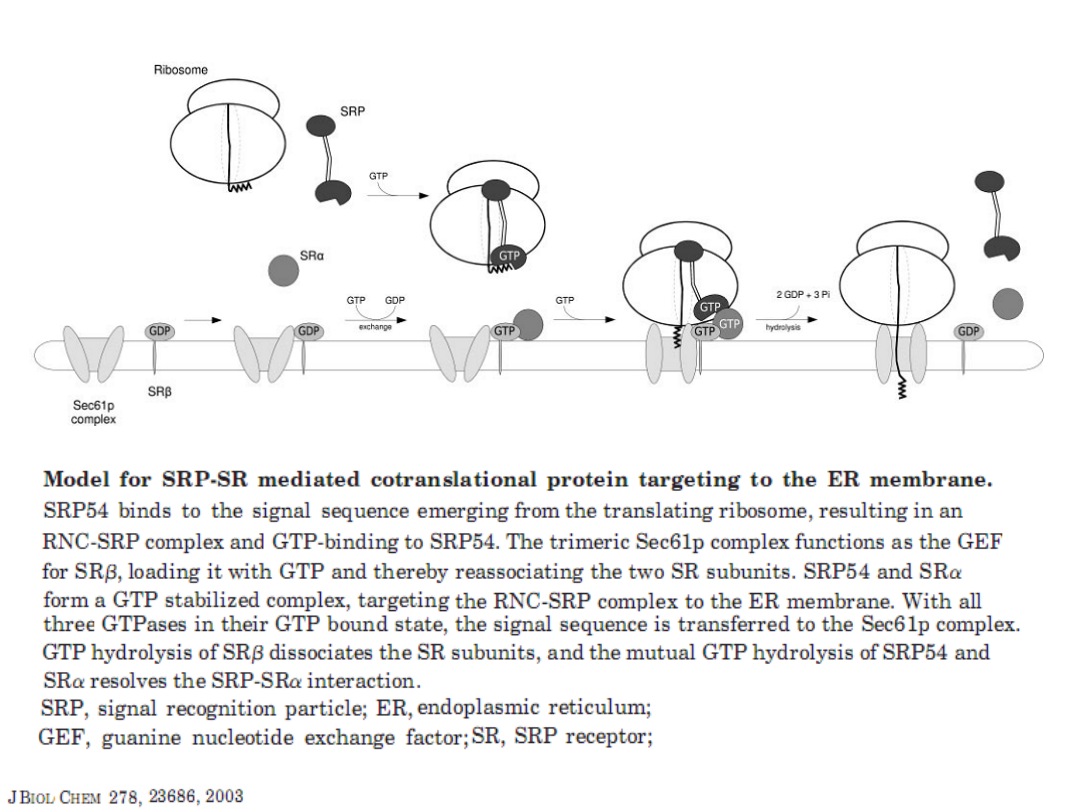
The model of the steps in eukaryotic translation
initiation
and the roles of the factors.
Annu.Rev.Biochem. 73:657, 2004
Annu.Rev.Biochem. 73:657, 2004
The model for translation elongation in eukaryotes
Annu.Rev.Biochem. 73:657, 2004
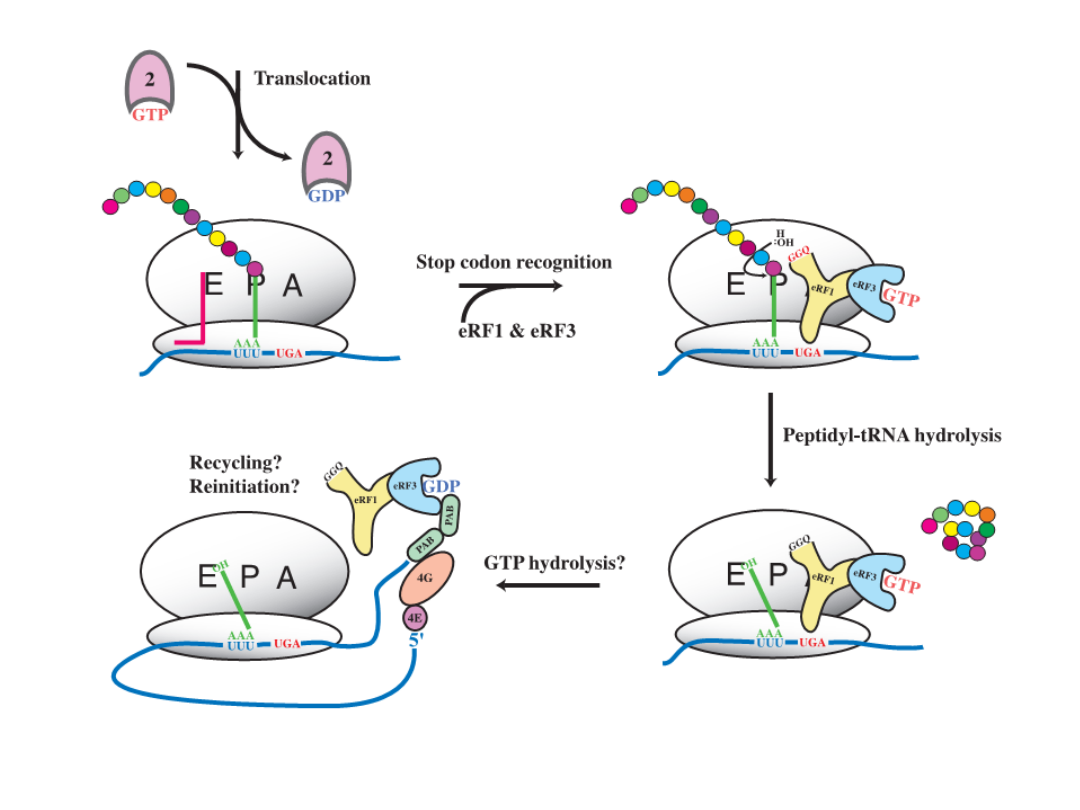
The model for translation termination in eukaryotes
mod.: Molecular Cell 28, 721,
2007
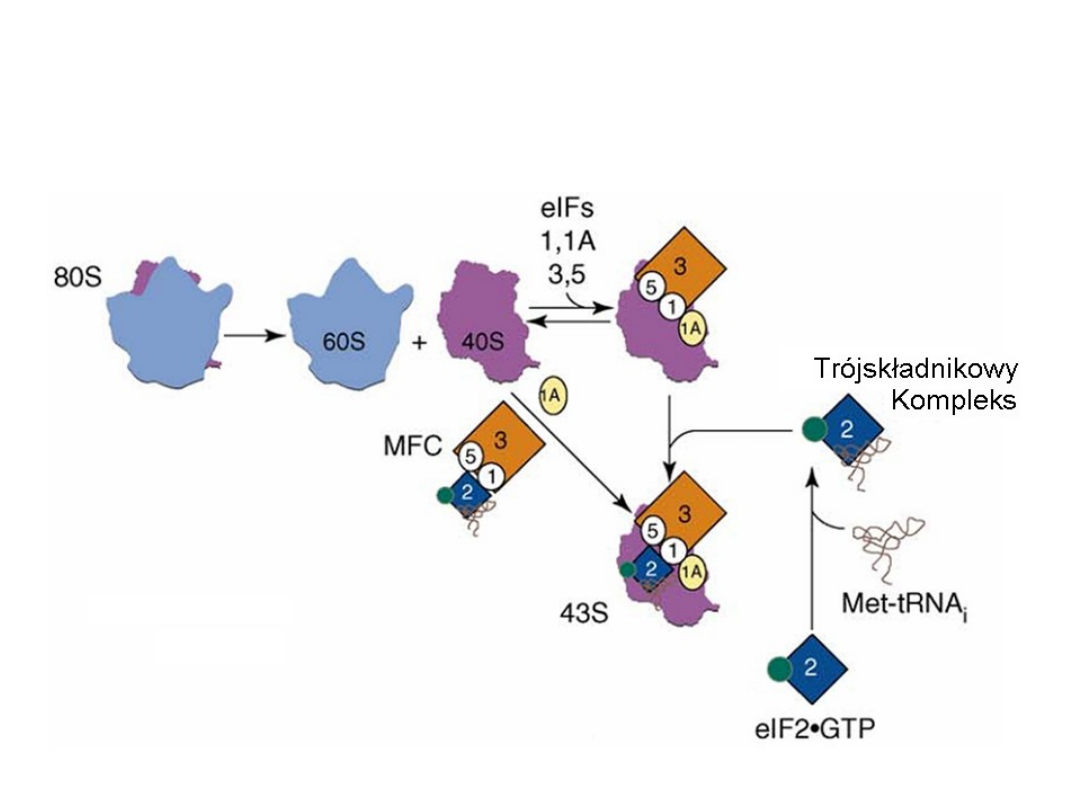
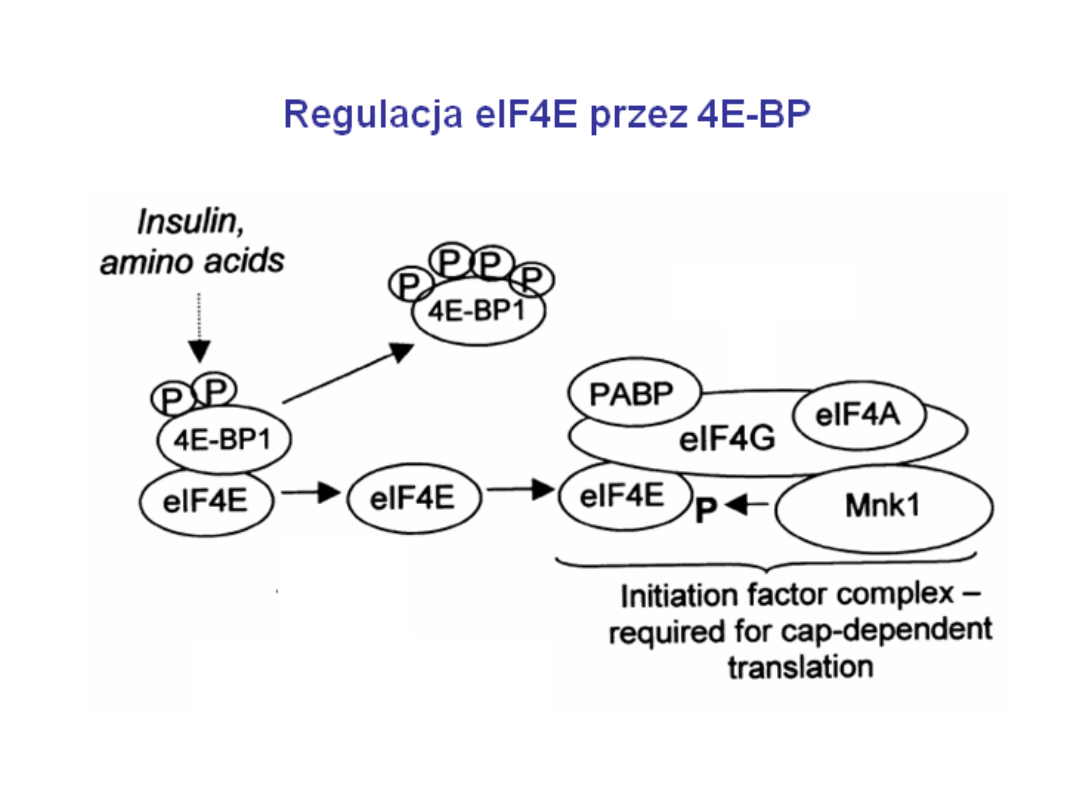
Inicjacja translacji w komórkach eukariotycznych
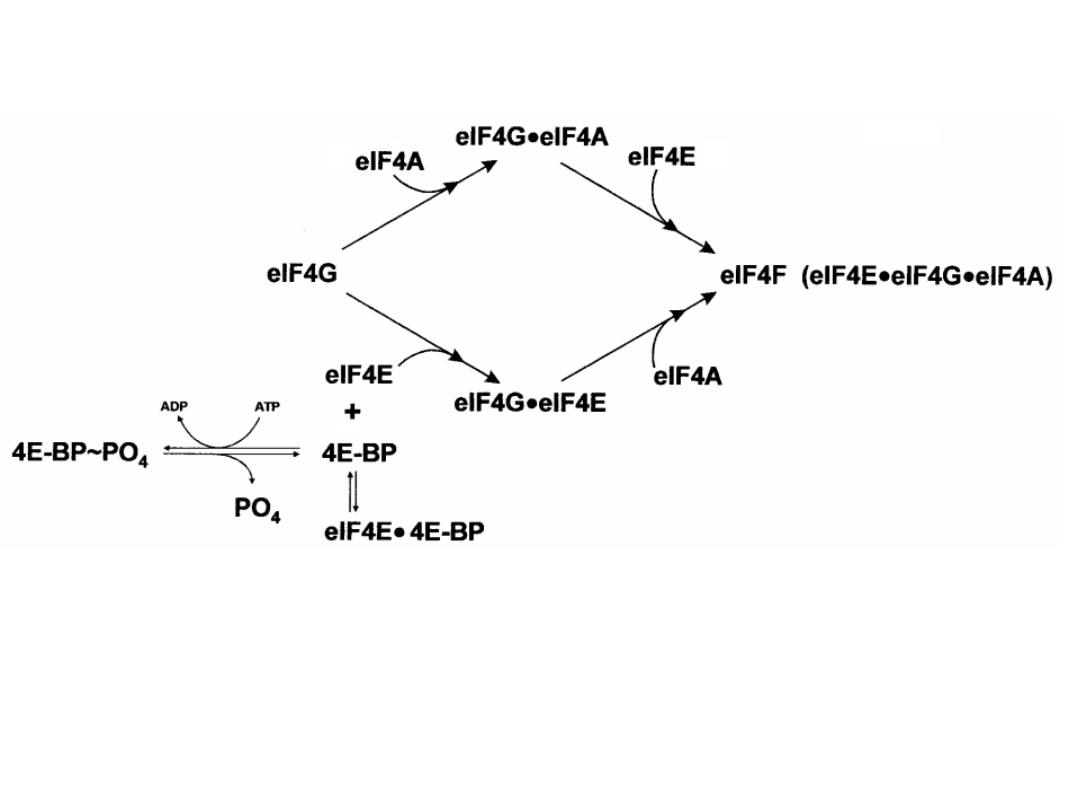
Regulation of eIF4F activity via the sequestration of eIF4E by 4E-BP.
The protein kinases that regulate the phosphorylation state of 4E-BP
emanate from them TOR and S6 kinase pathways. In general, these
pathways are activated by conditions that favor growth and are
inactivated by stress conditions.
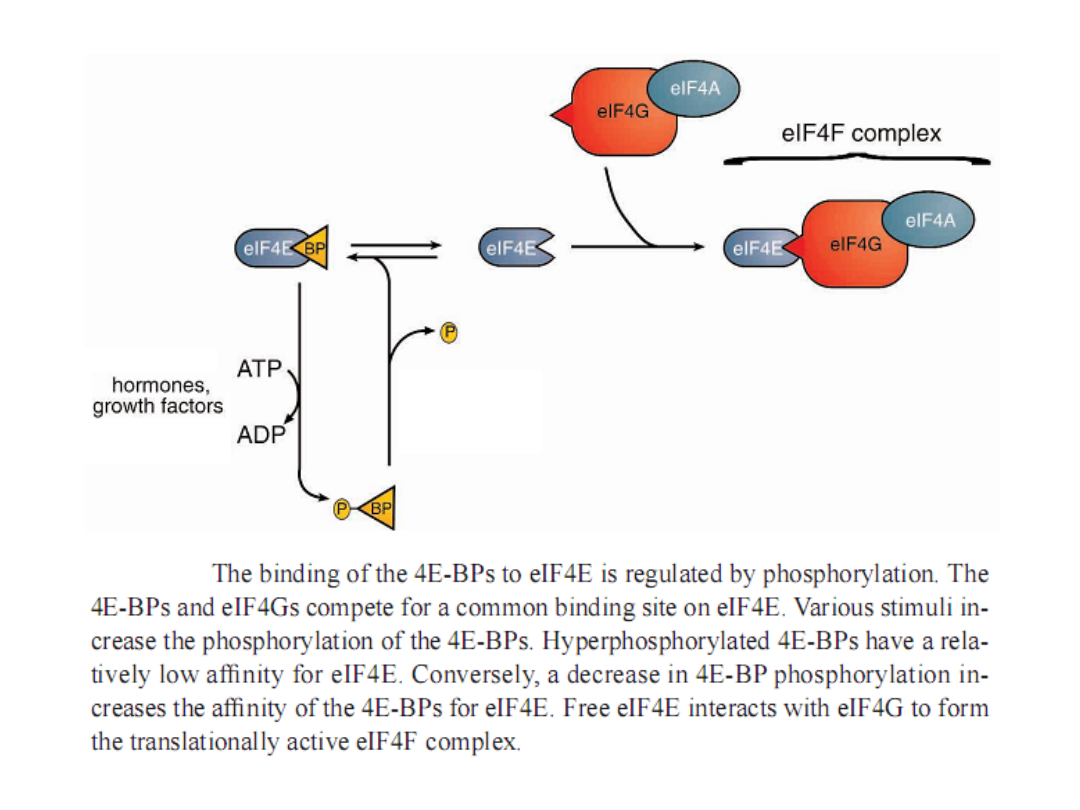
Annu. Rev. Biochem. 68:913, 1999
Genes & Dev. 15: 1593,
2001
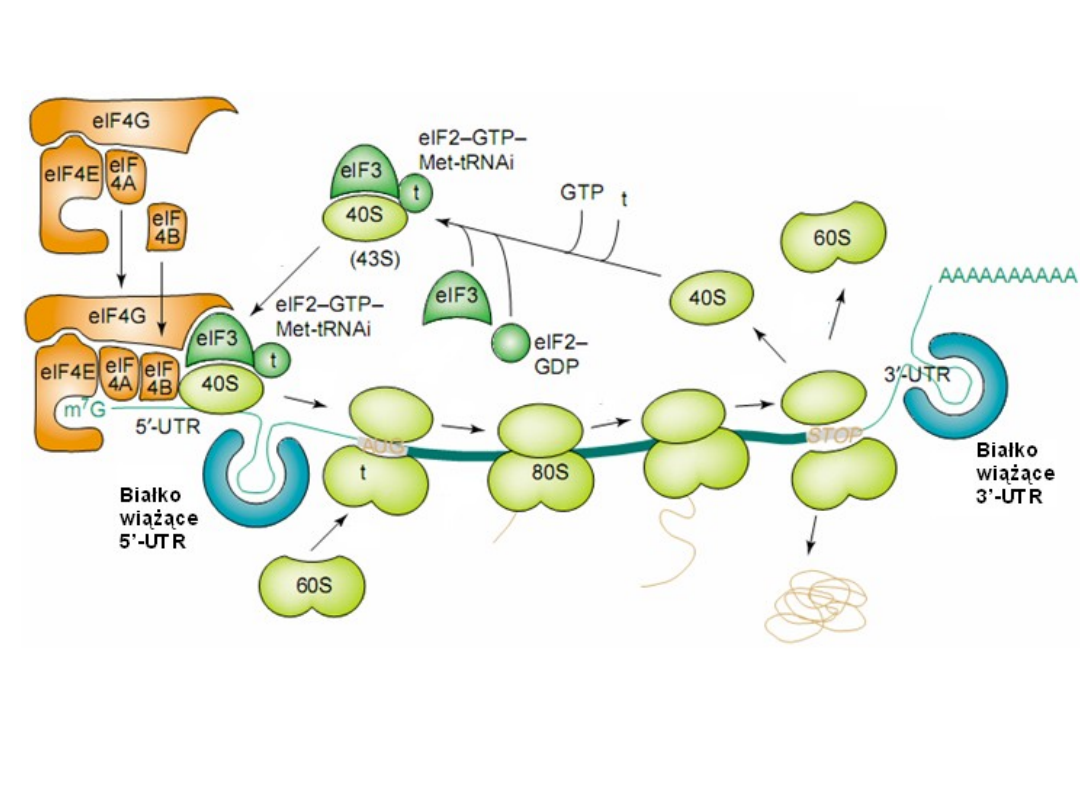
Classical protein translation and mechanisms
of control.
TRENDS in Biochemical Sciences 28, 91,2003
5'- UTR = 5'-untranslated region
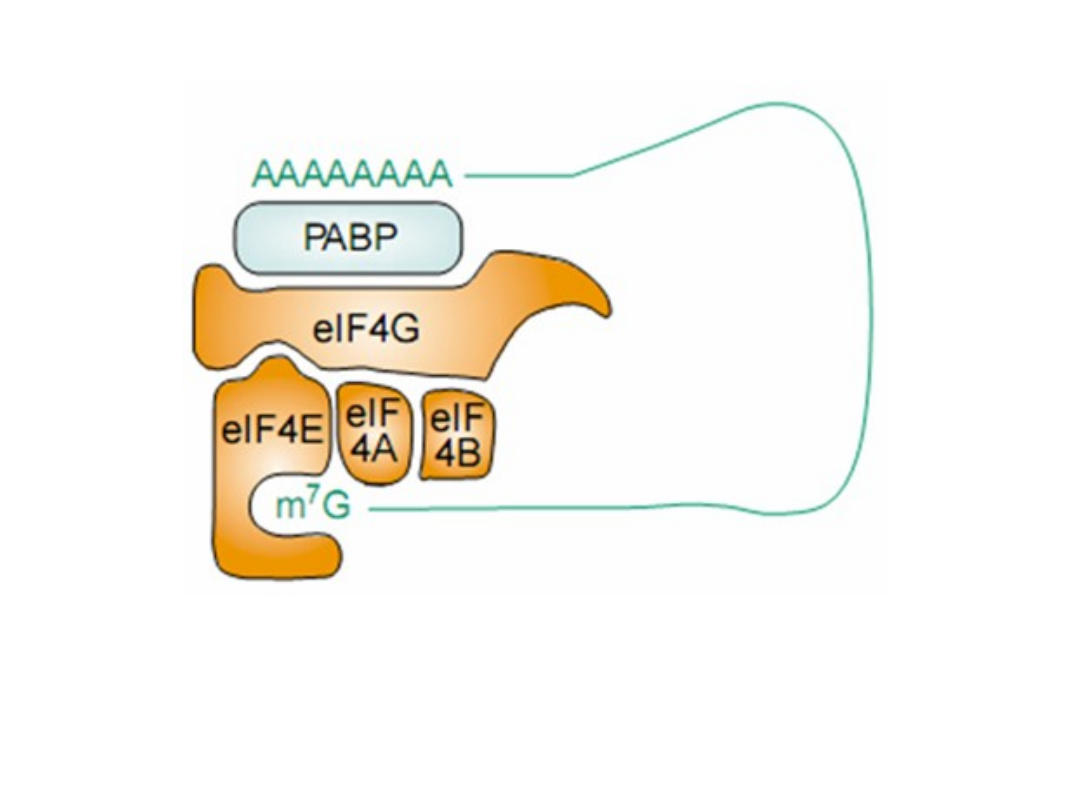
TRENDS in Biochemical Sciences 28, 91,2003
Multiple types of end-to-end interactions of RNA. Poly(A)-binding
protein (PABP)-mediated interaction of eukaryotic intiation factor
(eIF) 4G and the poly(A) tail in capped, polyadenylated human
mRNA.
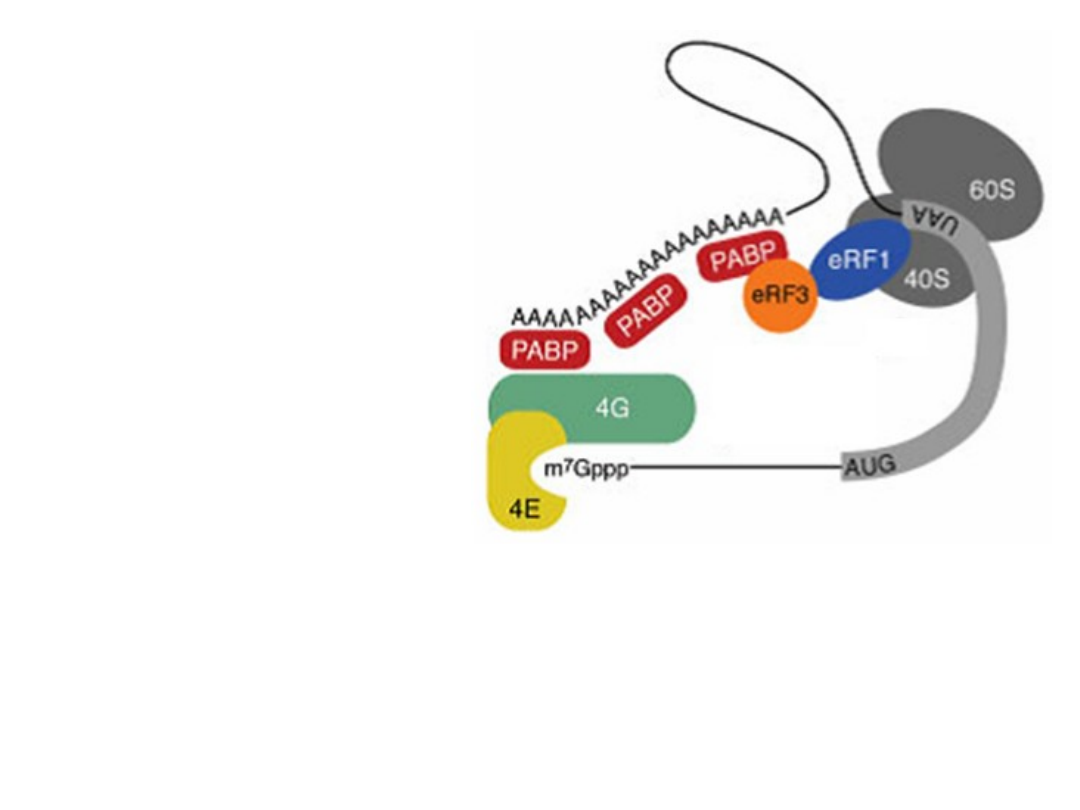
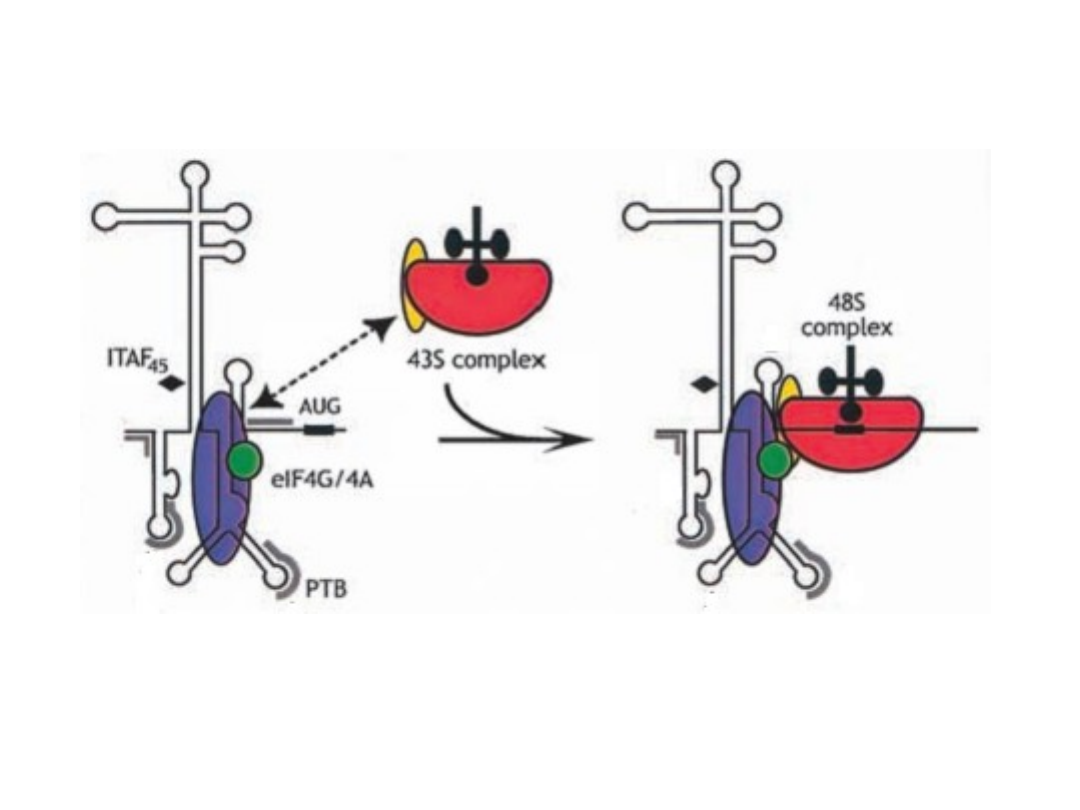
The ribosome - recycling concept.
A possible function of mRNA circularization could in volve facilitation of a
direct recycling of ribosomes or ribosomal subunits, after termination at
the stop codon, back to the 5’ region of the same mRNA (indicated by
the green arrow). This speculative model is supported by the
observation of circular polyribosomes, as well as interactions between
PABP, initiation factors bound to the cap structure, and the translation
termination factore eRF3.
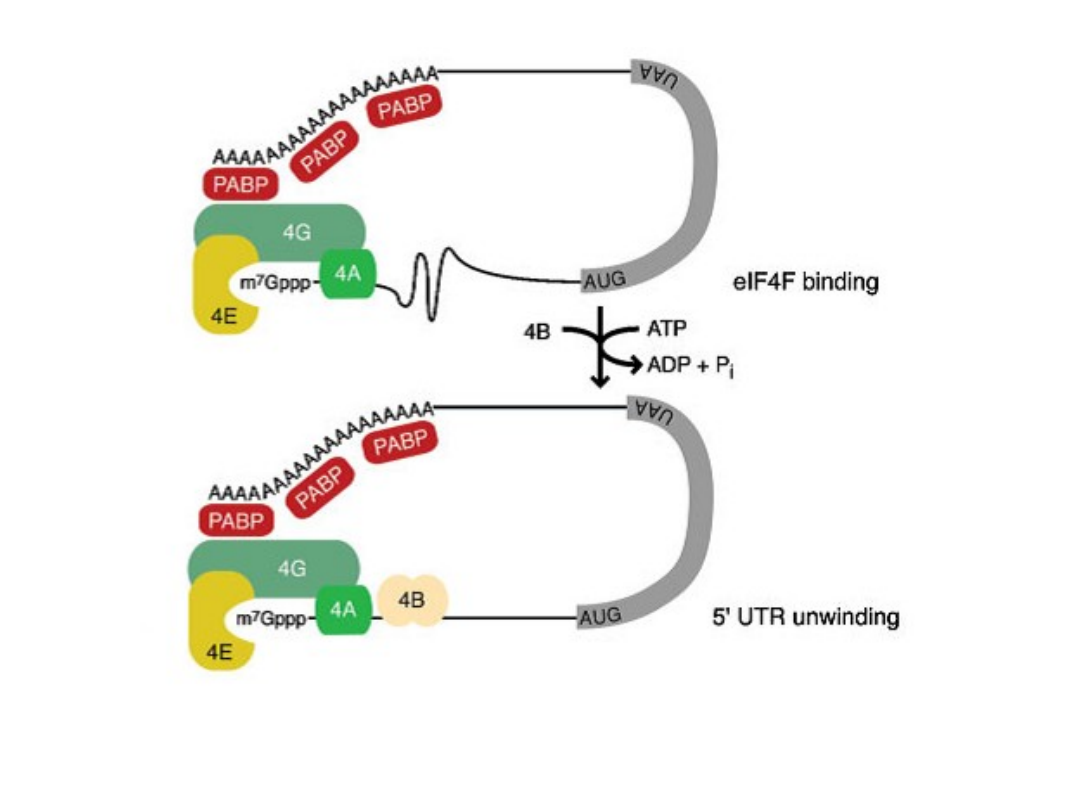
TIBS 28, 91, 2003
UTR = 5’- untranslated region
TIBS 28, 91, 2003
ORF (Open Reading Frame) - Otwarta ramka odczytu
Protein maturation: modification, secretion, targeting
5’
AUG
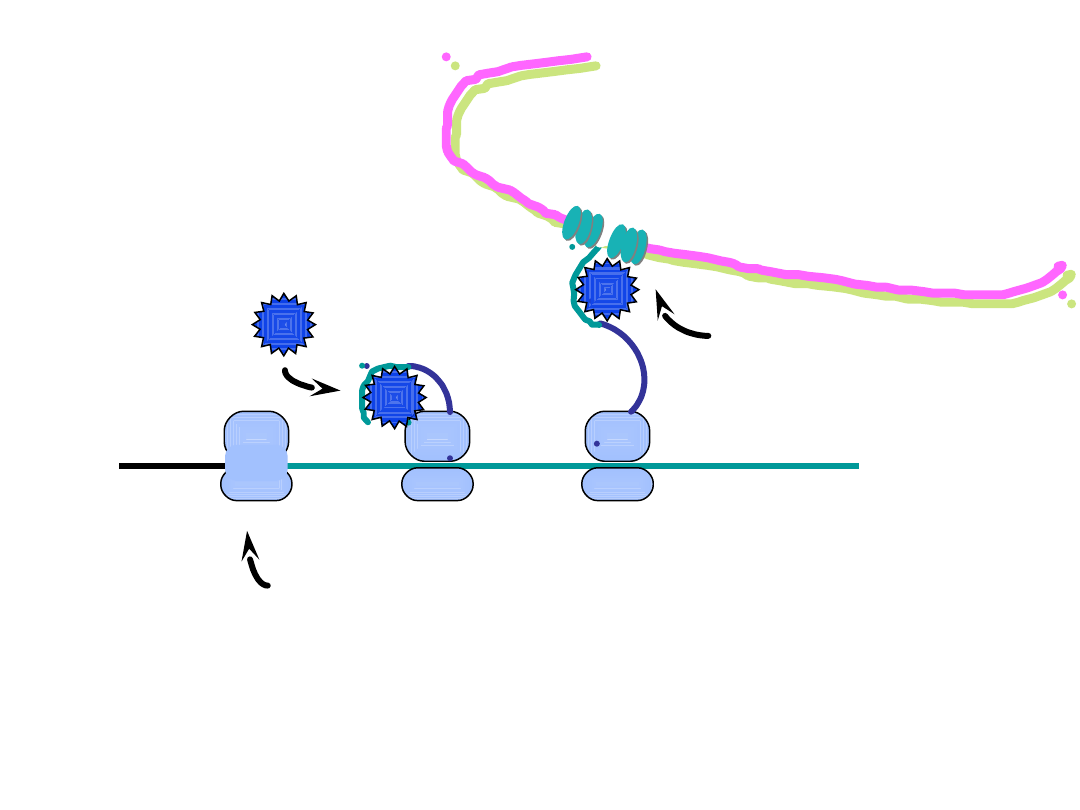
polysome for secreted protein
2. the signal recognition particle
a
(SRP)
binds the signal peptide
b
and
halts translation
1. translation initiates as usual
on a cytosolic mRNA
a
the
signal recognition particle (SRP)
consists of protein and RNA (7SL RNA); it binds
to the signal peptide, to the ribosome, and to the SRP receptor on the ER membrane
b
the signal peptide is a polypeptide extension of 10-40 residues, usually at the N-terminus
of a protein, that consists mostly of hydrophobic amino acids
c
ER = endoplasmic reticulum
ER
lumen
c
cytosol
3. the SRP docks with the SRP receptor on
the cytosolic side of the ER membrane
and positions the signal peptide for
insertion through a pore
SRP
SRP receptor
Translation of a secreted protein
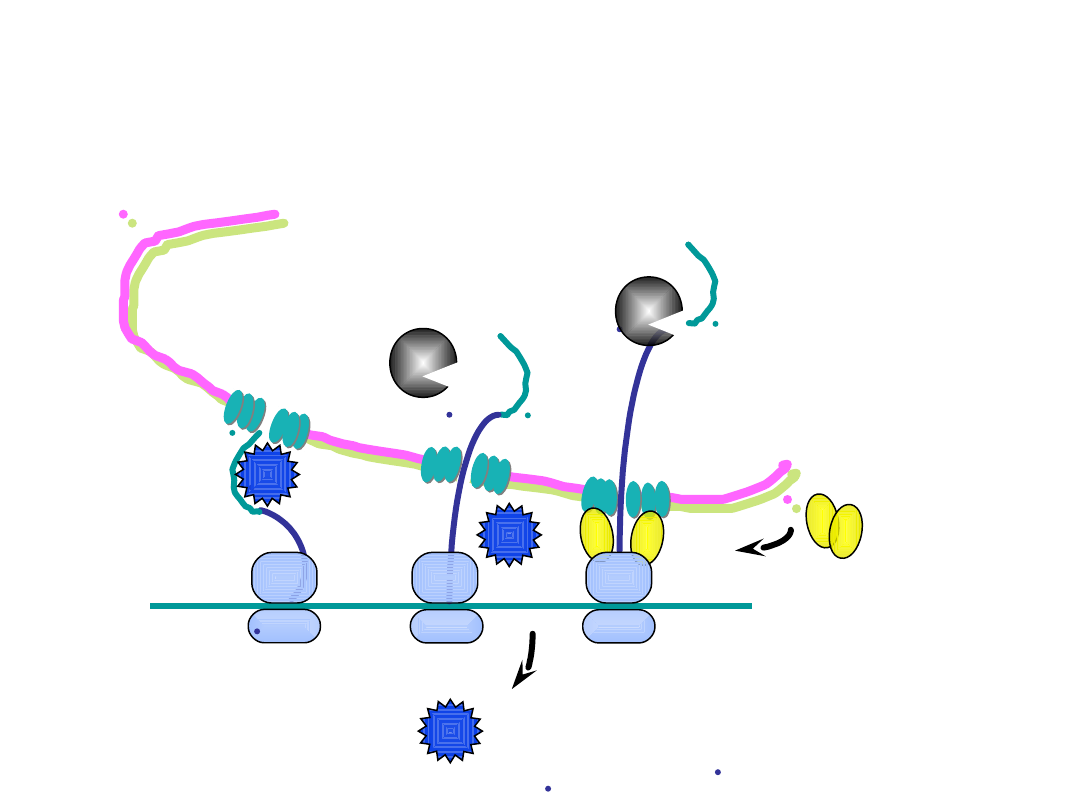
5’
ER lumen
cytosol
4. translation resumes and the nascent
polypeptide moves into the ER lumen
5.
signal peptidase
, which is in the ER
lumen, cleaves off the signal peptide
7. the ribosomes dock onto the
ER membrane; the rough ER
is ER studded with polysomes
6. the SRP is released
and is recycled
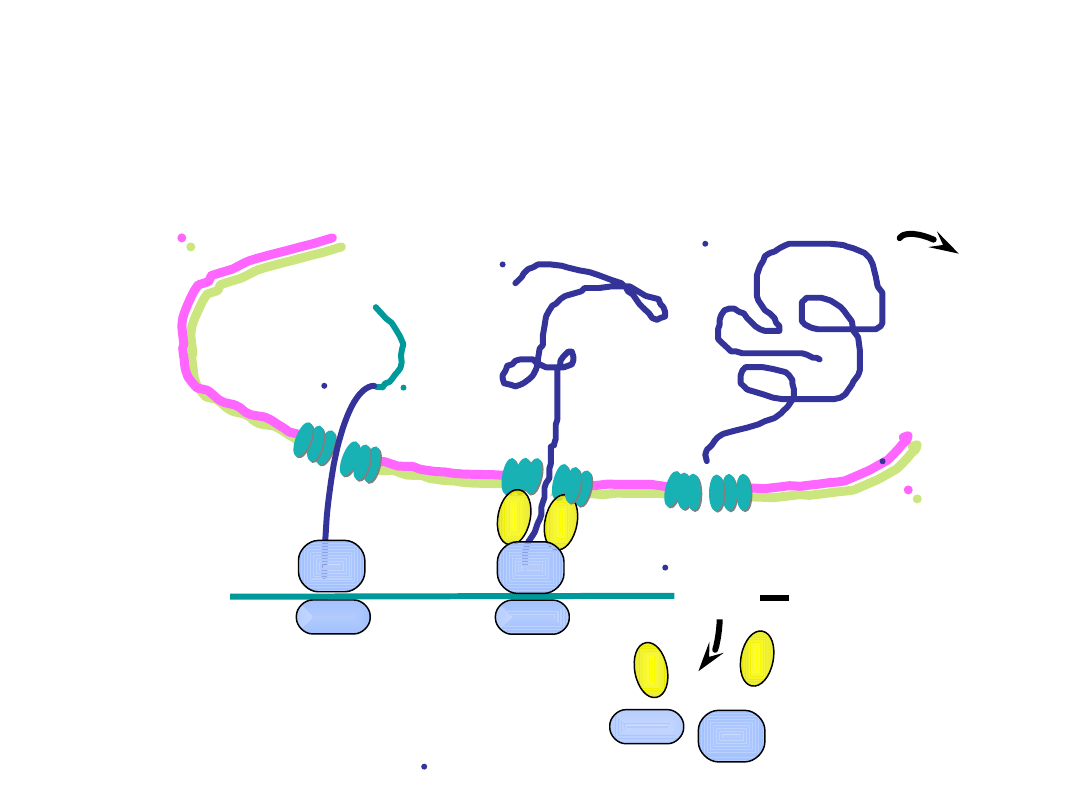
5’
ER lumen
cytosol
UGA
8. translation continues with the nascent
polypeptide emerging into the ER lumen
9. at termination of translation, the completed protein is
within the ER and is further processed prior to secretion
completed
protein is
processed and
secreted
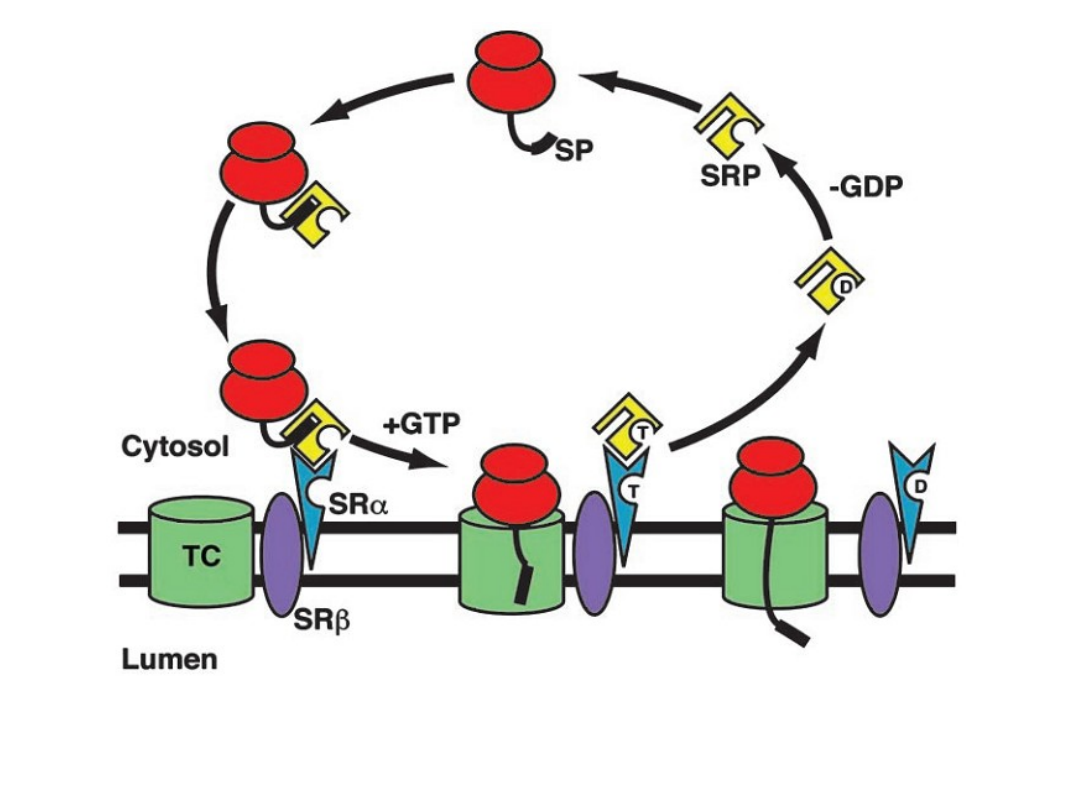
Annu.Rev.Biochem. 73:539, 2004

• Examples of secreted proteins:
• polypeptide hormones (e.g., insulin)
• albumin
• collagen
• immunoglobulins
• Integral membrane proteins are also synthesized by the same mechanisms;
they may be considered “partially secreted”
• Examples of integral membrane proteins:
• polypeptide hormone receptors (e.g., insulin receptor)
• transport proteins
• ion channels
• cytoskeletal anchoring proteins (e.g., band 3)

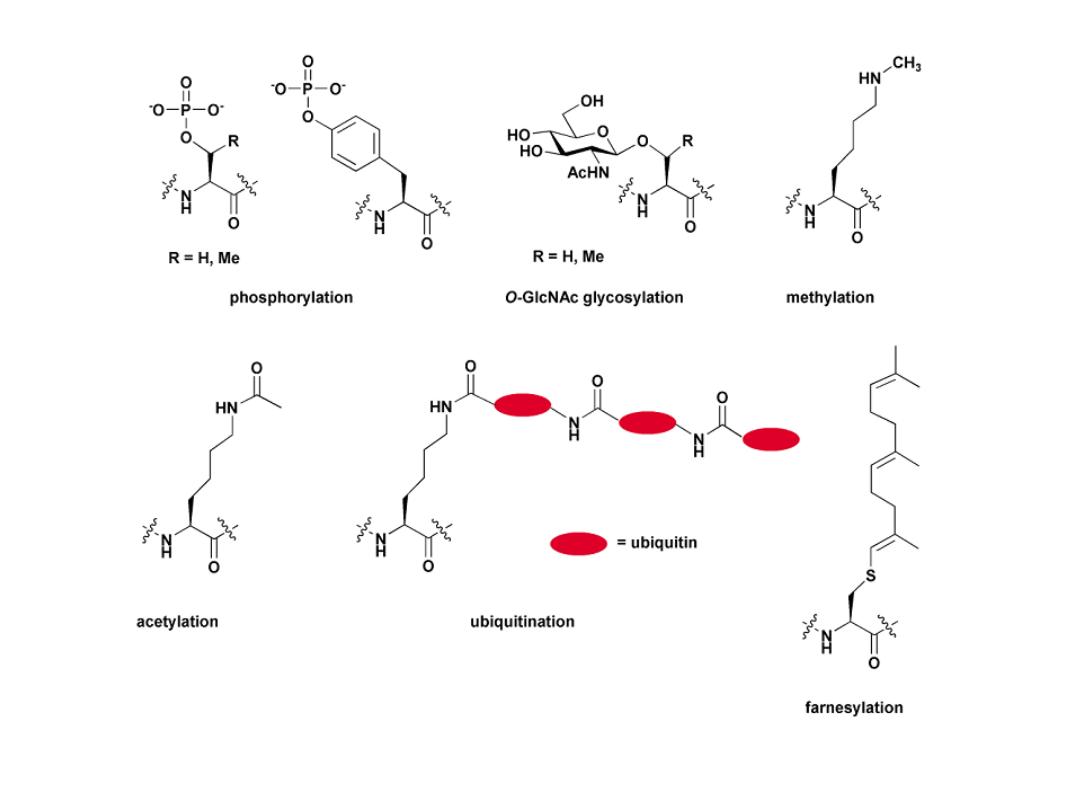
Some examples of posttranslational modications.
Org. Biomol. Chem.2, 1, 2004


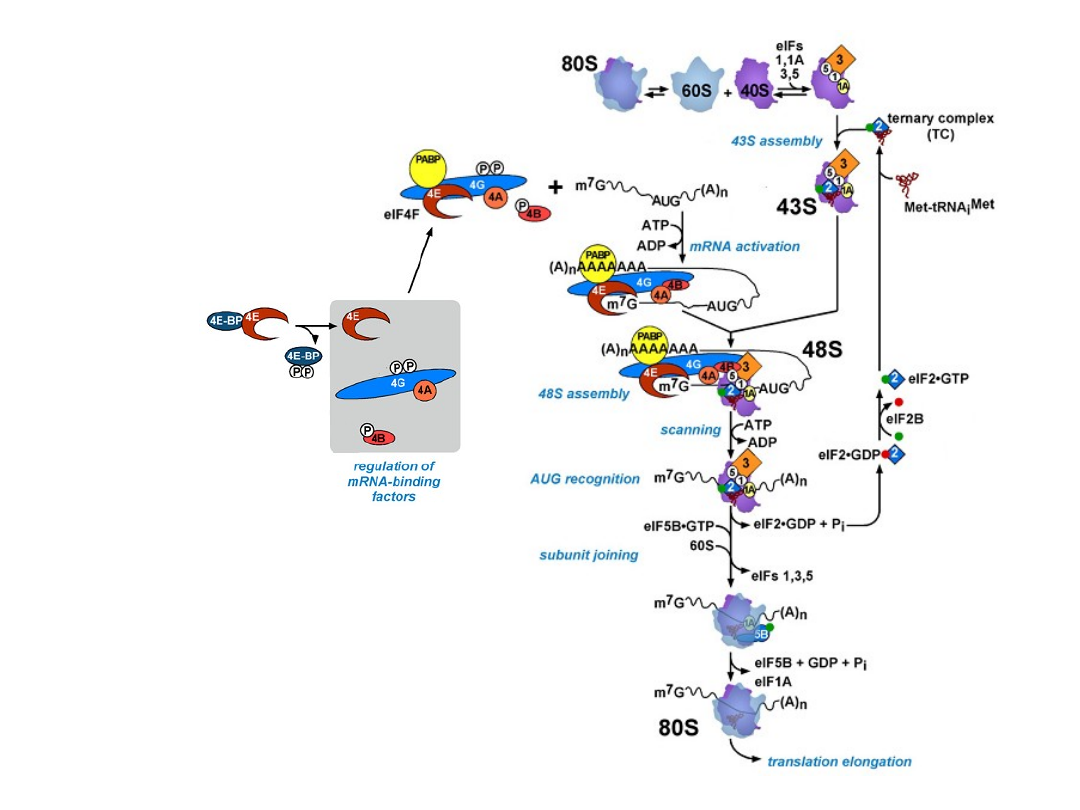
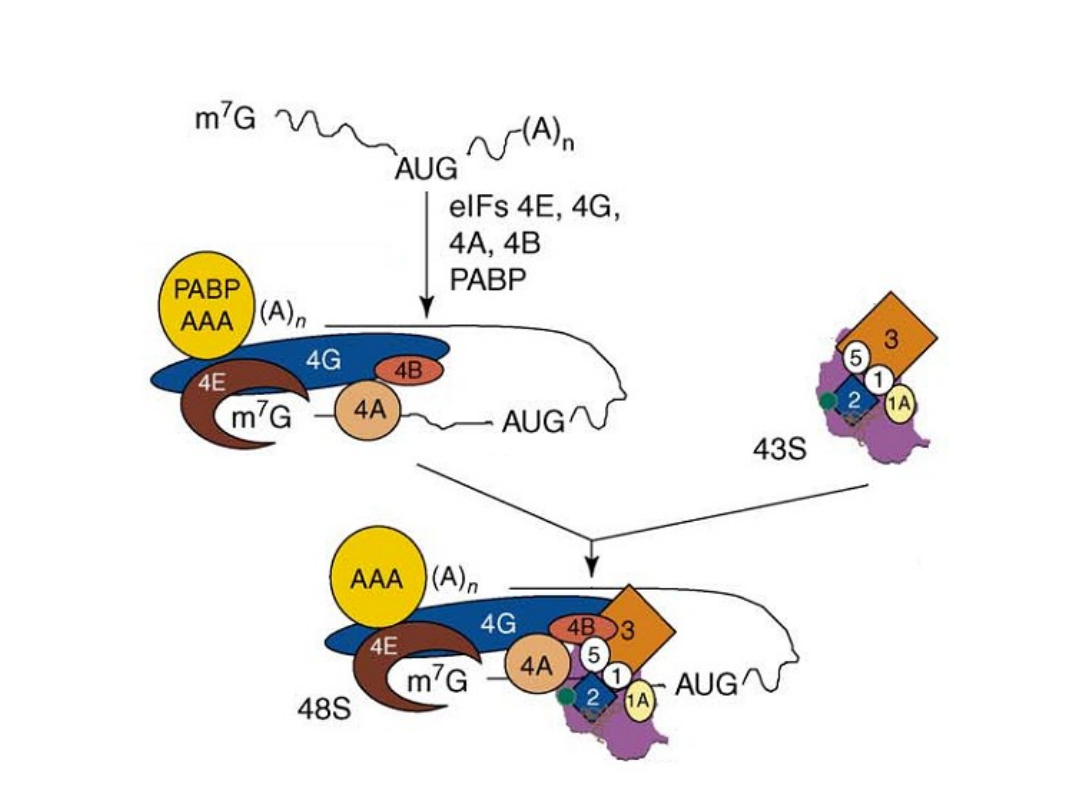
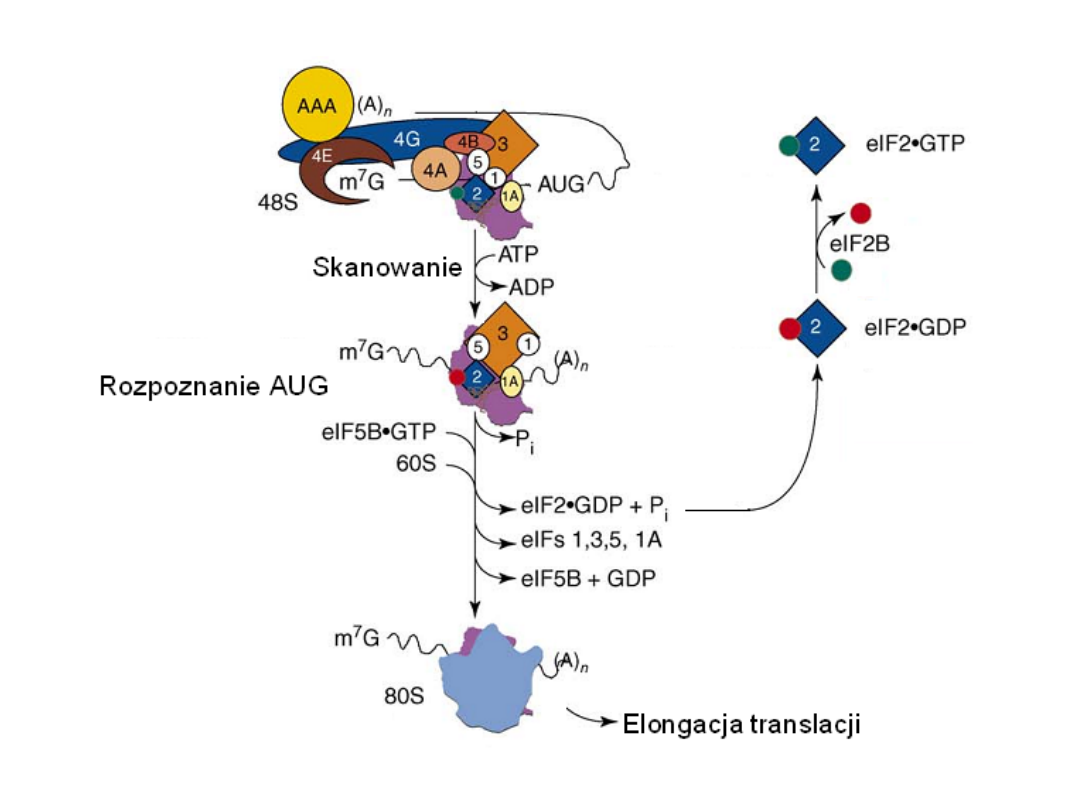
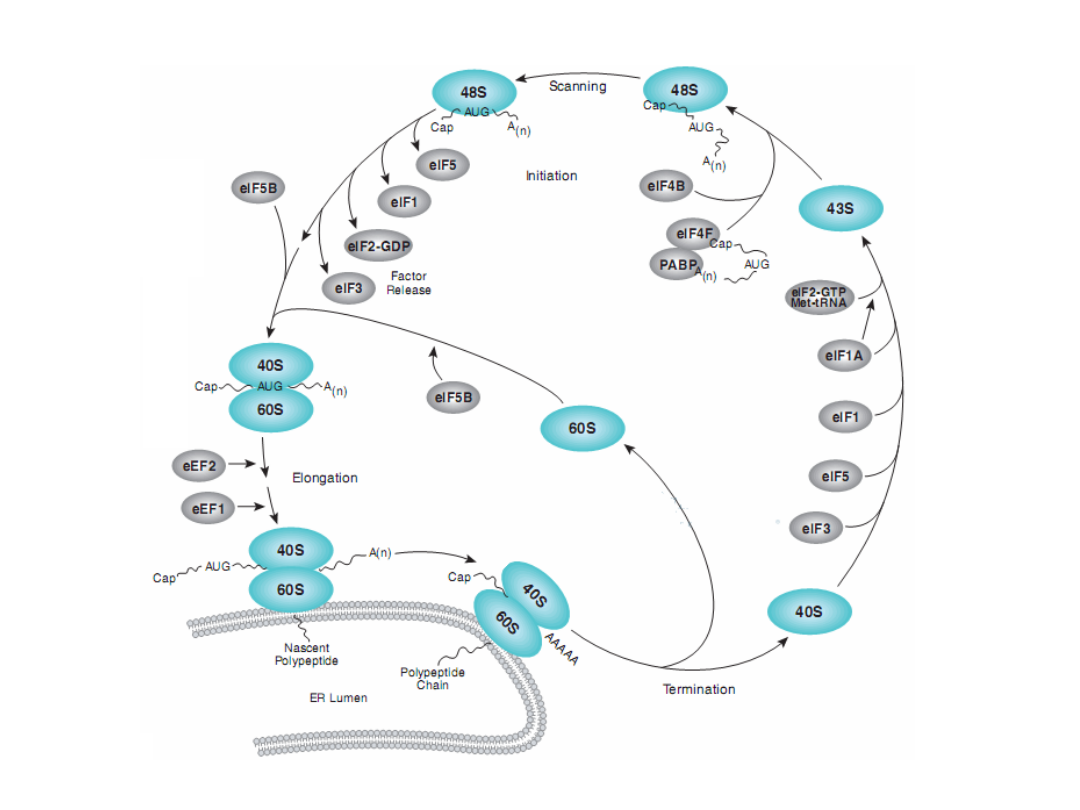
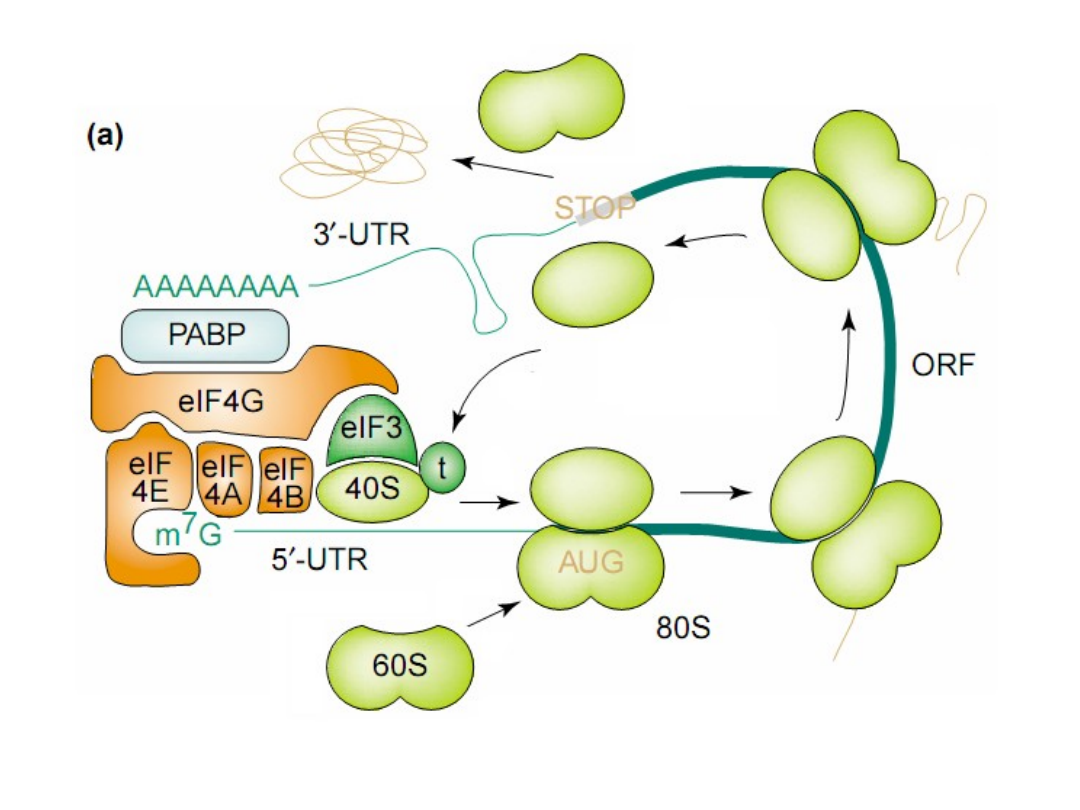
Classical protein translation and mechanisms of control. (a) Major events
in translation-initiation are: (i) the
eIF4F initiation complex, consisting of
eIF4A, eIF4G, and eIF4E
to the m7GpppN cap, is recruited and is joined by
eIF4B; (ii) the
48S ribosomal initiation complex
is formed by the joining of
the 43S pre-initiation complex, which contains eIF3, the 40S ribosomal
subunit and eIF2-GTP-Met-tRNAi, (iii) the 48S complex scans across the 5’-
UTR to the AUG initiation codon; and (iv) AUG is recognized by the
anticodon in Met-tRNAi and the 60S ribosomal subunit is recruited, which
is accompanied by hydrolysis of eIF2-bound GTP and dissociation of
initiation factors, to form a translation-competent 80S ribosome. during
protein elongation (v-vii), multiple 80S ribosomes simultaneously transit
the open reading frame (ORF) while elongating the newly synthesized
peptide chain [which requires aminoacid-charged tRNAs and elongation
factors (notshown)]. During termination (viii), the ribosome recognizes a
stop codon and the 80S ribosomal subunits dissociate from the mRNA in a
process requiring release factors (not shown). Also shown are
5’- and 3’-
untranslated region (UTR)-binding proteins
.
TIBS 28, 91, 2003

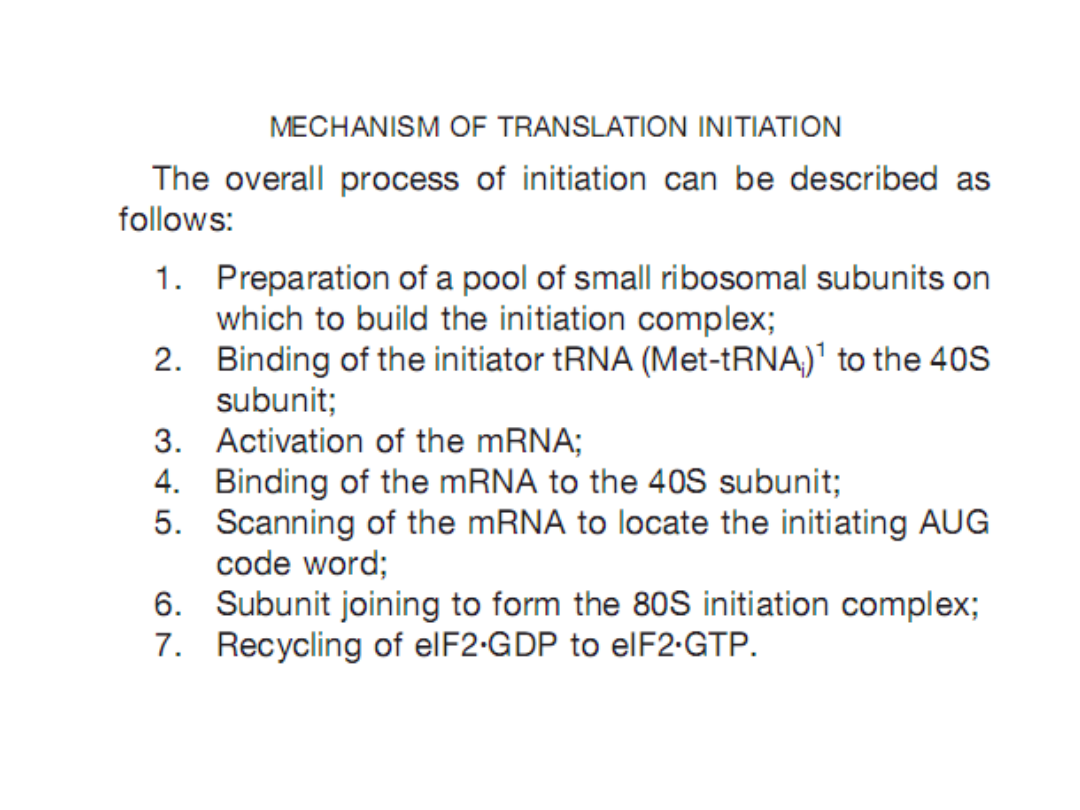
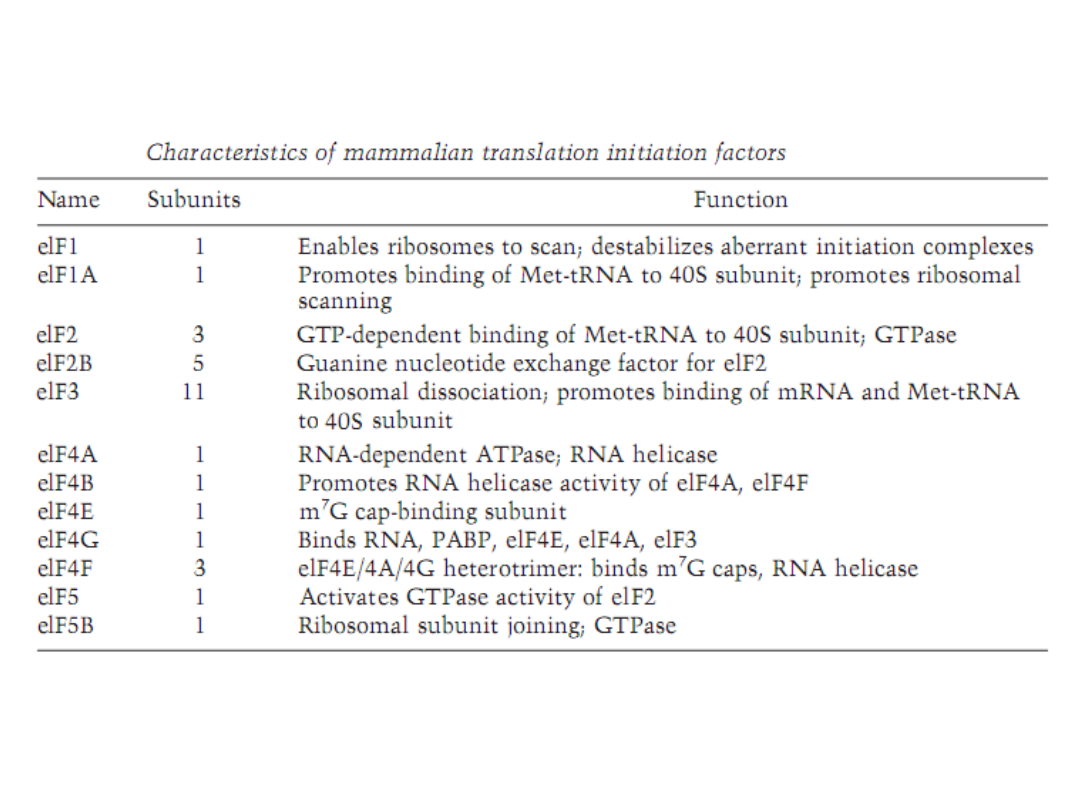
Initiation of protein synthesis in eukaryotesis important both
mechanically, because it selects the translation reading frame, and
biologically, because It is the primary site for regulation of
translation. For approximately 95–98% of the cellular mRNAs,
formation of an initiation complex follows a rather specific pathway
that is broken down in to seven discrete steps.
Although almost 35
peptids in12–14 translation initiation factors participate in this
process, three appear to have dominant roles: eukaryotic initiation
factor (
eIF
)
3
, in building a pool of 40S subunits;
eIF2
, in binding the
initiator tRNA (tRNAi) to the 40S subunit; and
eIF4F
inactivating the
mRNA and binding it to the 40S subunit.
This process requires both
ATP and GTP. There sulting 80S initiation complex contains both the
tRNAi and them RNA, with the anticodon of the tRNAi correctly base
paired with the initiating AUG code word. Regulation of translation
focuses mostly on controlling the activity of either eIF2 or eIF4F and
this regulation has different consequences. Reduction in eIF2 activity
influences all mRNA approximately the same, where as reductionin
eIF4F activity drives competition between mRNAs.
BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION 31, 378, 2003
Initiation of Protein Biosynthesis in Eukaryotes
Document Outline
- Slide 1
- Slide 2
- Slide 3
- Slide 4
- Slide 5
- Slide 6
- Slide 7
- Slide 8
- Slide 9
- Slide 10
- Slide 11
- Slide 12
- Slide 13
- Slide 14
- Slide 15
- Slide 16
- Slide 17
- Slide 18
- Slide 19
- Slide 20
- Slide 21
- Slide 22
- Slide 23
- Slide 24
- Slide 25
- Slide 26
- Slide 27
- Slide 28
- Slide 29
- Slide 30
- Slide 31
- Slide 32
- Slide 33
- Slide 34
- Slide 35
- Slide 36
- Slide 37
- Slide 38
- Slide 39
- Slide 40
- Slide 41
- Slide 42
- Slide 43
- Slide 44
- Slide 45
- Slide 46
- Slide 47
- Slide 48
- Slide 49
- Slide 50
- Slide 51
- Slide 52
- Slide 53
- Slide 54
Wyszukiwarka
Podobne podstrony:
wykład 08 - pedagogika behawioralna - Winfired Wermter - Dom Mi, współczesne kierunki pedagogiczne
MT I Wyklad 08
MC W Wyklad 08 Tlenkowe Materialy Konstrukcyjne
26) TSiP Wyklad 08 pekanie
fiz wyklad 08
krajoznawstwo, wykład I 08.10.2007, CIASTO NA NALEŚNIKI
Wykład 08.05.2010
Wykład 08, 05
Teoria Informacji Wykład 6 (08 04 2015)
B. W. w Unii Europejskiej - wyklad 08.10, Sudia - Bezpieczeństwo Wewnętrzne, Semestr III, Bezpieczeń
Encyklopedia Prawa - wyklad 08 [06.11.2001], INNE KIERUNKI, prawo, ENCYKLOPEDIA PRAWA
2006C16 wyklad 08 (2)
Ekologiczne Systemy Chowu i Żywienia Zwierząt - Wykład 08, WYKŁAD VIII- EKOLOGICZNE SYSTEMY CHOWU I
Powszechna historia prawa - wykłady, 08.01.2013
Powszechna historia prawa - wykłady, 08.01.2013
32) TSiP Wyklad 08 plastycznosc
Algebra I wyklad 08
materiały do wykładów, w 08 Szkoła jako instytucja społeczna i środowisko wychowawcze
więcej podobnych podstron