
MYB96-mediated abscisic acid signals induce pathogen
resistance response by promoting salicylic acid
biosynthesis in Arabidopsis
Pil Joon Seo
1
and Chung-Mo Park
1,2
1
Department of Chemistry, Seoul National University, Seoul 151-742, Korea;
2
Plant Genomics and Breeding Institute, Seoul National University, Seoul
151-742, Korea
Author for correspondence:
Chung-Mo Park
Tel: +82 (2) 880 6640
Email: cmpark@snu.ac.kr
Received: 30 October 2009
Accepted: 13 December 2009
New Phytologist (2010)
186: 471–483
doi: 10.1111/j.1469-8137.2010.03183.x
Key words: abscisic acid, Arabidopsis,
disease resistance, MYB96, salicylic acid,
SID2.
Summary
• The Arabidopsis MYB96 transcription factor plays a role in abscisic acid (ABA)-
mediated drought response. Notably, anthocyanins accumulate in the activation-
tagging myb96-1d line, suggesting a role of MYB96 in biotic and abiotic stress
responses in plants. Here, we investigate the role of MYB96 in salicylic acid (SA)
biosynthesis and plant defense and explore the mechanisms underlying the ABA–
SA interaction.
• myb96-1d and myb96-1 were subject to pathogen infection assays, and
expression of SA biosynthetic and defense genes was examined. myb96-1d was
crossed with the NahG transgenic plants to investigate the role of MYB96 in ABA
regulation of SA biosynthesis.
• Whereas myb96-1d exhibited an enhanced disease resistance, myb96-1 was sus-
ceptible to pathogen infection. A subset of pathogenesis-related (PR) genes was
up-regulated in myb96-1d. However, PR transcript abundances were reduced in
myb96-1d X NahG. Interestingly, a SA biosynthetic gene SALICYLIC ACID
INDUCTION DEFICIENT2 (SID2) was up-regulated, and concentrations of SA and
SA-b-glucoside (SAG) were elevated in myb96-1d. In addition, the inductive
effects of abiotic stresses on SID2 were reduced in aba3-1.
• Our observations indicate that MYB96-mediated ABA signals enhance plant
disease resistance by inducing SA biosynthesis. It is envisioned that MYB96 is a
molecular link that mediates ABA-SA crosstalks.
Introduction
Plants adapt to environmental fluctuations by adjusting
their physiology and morphology. Numerous genes are reg-
ulated during plant responses to biotic and abiotic stress
conditions. With an aim of improving stress adaptability
and productivity of crop plants, intensive works have been
carried out to identity genes and molecular mechanisms
underlying plant adaptation under various stress conditions
(Ingram & Bartels, 1996; Schenk et al., 2000; Seki et al.,
2002; Tao et al., 2003).
The stress genes function in a coordinate manner through
a complex signaling network as well as through individual
signaling pathways. While initial stimuli are obviously
diverse, these signals are integrated into a unified scheme in
many cases, resulting in common plant responses to different
stress signals (Albrecht et al., 2003; Denekamp & Smeekens,
2003; Park et al., 2007). An example is a group of pathogene-
sis-related (PR) genes. They are well-known marker genes for
plant pathogenesis that play primary roles in disease resis-
tance response (Bol et al., 1990). Notably, it has been
recently reported that the PR genes are also induced by abi-
otic stresses, such as cold, high salinity, and drought (Seo
et al., 2008). Abiotic stresses are also known to confer disease
resistance in Arabidopsis (Gaudet et al., 2003; Griffith &
Yaish, 2004). An activation tagging allele of the Activated
Disease Resistance 1 (ADR1) gene encoding the coiled-coil
(CC) nucleotide-binding site (NBS) leucine-rich repeat
(LRR) protein exhibits drought resistance as well as salicylic
acid (SA)-mediated resistance to virulent pathogens (Grant
New
Phytologist
Research
The Authors (2010)
Journal compilation New Phytologist Trust (2010)
New Phytologist (2010) 186: 471–483
471
www.newphytologist.com

et al., 2003; Chini et al., 2004), supporting a wide range of
signaling crosstalks between biotic and abiotic stress signals.
Salicylic acid is an important growth hormone functioning
in plant–microbe interactions. Impaired SA biosynthetic
mutants, such as salicyclic acid induction deficient2 (sid2), and
NahG transgenic plants overexpressing a salicylate hydroxy-
lase that prevents accumulation of SA exhibit an increased
susceptibility to pathogen infection by compromising the
establishment of systemic acquired resistance (Gaffney et al.,
1993; Wildermuth et al., 2001). SA also plays regulatory
roles in plant response to various abiotic stresses. SA-deficient
NahG transgenic plants are resistant to oxidative damage
generated by osmotic stress (Borsani et al., 2001), although it
is currently unclear whether the resistance is attributable to
the reduced SA content or to the accumulated catechol in the
transgenic plants (Borsani et al., 2001). In addition, the
inhibitory effect of high salt and osmotic stress on seed germi-
nation is compromised by gibberellic acid (GA)-mediated
induction of SA biosynthesis (Alonso-Ramı´rez et al., 2009).
Salicylic acid is mainly synthesized through the isochoris-
mate pathway (Wildermuth et al., 2001; Garcion et al.,
2008). Although the biochemical activity is not fully char-
acterized, the SID2 gene plays a central role in the SA bio-
synthetic pathway (Wildermuth et al., 2001). SA is also
synthesized from phenylalanine by phenylalanine ammonia
lyase (PAL) activity (Lee et al., 1995), although its contri-
bution to endogenous SA content is relatively lower than
that of the isochorismate pathway.
While SA biosynthetic pathway genes have been relatively
well characterized, regulatory mechanisms governing SA
biosynthesis are poorly understood: only a few transcription
factor genes have been reported to regulate SA biosynthesis.
The Arabidopsis MYB30 transcription factor is related to
the hypersensitive cell death program (Raffaele et al.,
2006). The R2R3-type MYB transcription factor regulates
hypersensitive response by modulating SA accumulation.
Consequently, the MYB30-mediated cell death is abolished
in SA biosynthetic mutants but is unaffected in SA signaling
mutants, such as npr1 (Raffaele et al., 2006).
Anthocyanin accumulation is a prominent developmental
appearance that is caused by diverse environmental stresses,
such as ultraviolet light, nutrient deficiency, and abiotic
stress conditions (Winkel-Shirley, 2001, 2002). It is also
closely related to plant pathogenesis and frequently used as
an easily visible marker for plant disease resistance response
(Dixon, 2001). A protein complex composed of an MYB, a
basic helix-loop-helix (bHLH), and WD40 proteins, thus
designated an MBW complex, play a key role in anthocya-
nin biosynthesis (Broun, 2005; Koes et al., 2005). In Ara-
bidopsis, it has been observed that the TRANSPARENT
TESTA GLABRA1 (TTG1), which plays a central role in
constituting the complex, interacts with GLABRA3 (GL3),
ENHANCER OF GLABRA3 (EGL3), and TRANSPAR-
ENT TESTA8 (TT8) (Walker et al., 1999; Zhang et al.,
2003). However, it is currently unclear whether the GL3
and EGL3 proteins are components of the MBW complex.
In addition, two redundant MYB transcription factors,
PRODUCTION OF ANTHOCYANIN PIGMENT1
(PAP1) and PAP2, also participate in the flavonoid biosyn-
thetic pathway (Borevitz et al., 2000; Teng et al., 2005).
Recently, some of these genes have been reported to be
environmentally regulated and provide resistance to envi-
ronmental stresses (Rowan et al., 2009).
The MYB transcription factors, one of the largest tran-
scription factor families in plants, regulate diverse develop-
mental processes and plant responses to environmental
stimuli (Stracke et al., 2001), such as cell fate determination
(Lee & Schiefelbein, 1999) and biotic and abiotic stresses
(Mengiste et al., 2003; Jung et al., 2008). The MYB96
transcription factor, a R2R3-type MYB member, has
recently been shown to serve as a positive regulator of
drought resistance response. It enhances plant resistance to
drought stress by inducing the RD22 gene (Seo et al.,
2009). An activation tagging line myb96-1d exhibits an
enhanced drought resistance with reduced lateral roots. By
contrast, the drought resistance response is significantly
reduced in the MYB96-deficient
myb96-1 mutant. Interest-
ingly, the MYB96 gene also mediates the auxin–ABA inter-
actions during lateral root development. The MYB96 gene
modulates abscisic acid (ABA)-mediated abiotic stress sig-
nals in inducing a small group of GH3 genes encoding IAA-
conjugating enzymes and contributes to maintenance of
endogenous IAA contents at an appropriate amount under
drought conditions (Seo et al., 2009).
Here, we report that the MYB96 transcription factor links
ABA-mediated abiotic stress signals with SA biosynthesis
and pathogen resistance response. While the myb96-1d acti-
vation tagging line exhibited an enhanced disease resistance,
the myb96-1 mutant was more susceptible to a virulent Pseu-
domonas syringae DC3000 strain. Consistent with this, the
SID2 gene was markedly up-regulated, and endogenous con-
centrations of free SA and SA-b-glucoside (SAG) were ele-
vated in myb96-1d. Interestingly, the myb96-1d phenotypes,
including impaired leaf development and dwarfed growth,
were suppressed in the myb96-1d X NahG genetic cross,
indicating that SA is closely linked with MYB96-mediated
ABA signaling. It is therefore proposed that the MYB96 gene
serves as a molecular knot that integrates ABA- and SA-med-
iated signals under environmental stress conditions.
Materials and Methods
Plant materials and growth conditions
All Arabidopsis thaliana (L.) Heynh. lines used were in the
Columbia background (Col-0), unless otherwise specified.
Plants were grown in a controlled culture room set at 22C
with a relative humidity of 60% under long-day conditions
472
Research
New
Phytologist
The Authors (2010)
Journal compilation New Phytologist Trust (2010)
New Phytologist (2010) 186: 471–483
www.newphytologist.com

(16 h light and 8 h dark), with white light illumination
(120 lmol photons m
)2
s
)1
) provided by fluorescent
FLR40D ⁄ A tubes (Osram, Seoul, Korea). The activation-
tagging line myb96-1d and the myb96-1 knockout mutant
have been previously described (Seo et al., 2009).
Transcript abundance analysis
Quantitative real-time RT-PCR (qRT-PCR) was employed
for measuring transcript abundances. Total RNA sample
preparation, reverse transcription, and qRT-PCR were car-
ried out based on the rules that have recently been proposed
by Udvardi et al. (2008) to ensure reproducible and accu-
rate measurements. Extraction of total RNA samples from
appropriate plant materials and qRT-PCR conditions have
been previously described (Kim et al., 2006). The RNA
samples were extensively pretreated with an RNAse-free
DNAse to eliminate any contaminating genomic DNA
before use. The PCR primers used are listed in Supporting
Information, Table S1.
Quantitative real-time RT-PCR was carried out in 96-well
blocks with an Applied Biosystems 7500 Real-Time PCR
System (Foster City, CA, USA) using the SYBR Green I mas-
ter mix in a volume of 25 ll. The PCR primers were
designed using the Primer Express Software installed into the
system. The two-step thermal cycling profile used was 15 s at
94C and 1 min at 68C. An eIF4A gene (At3g13920) was
included in the assays as an internal control for normalizing
the variations in cDNA amounts used (Gutierrez et al.,
2008). The qRT-PCR reactions were carried out in biologi-
cal triplicates and technical duplicates using RNA samples
extracted from three independent plant materials grown
under identical growth conditions. The comparative DDC
T
method was used to evaluate the relative quantities of each
amplified product in the samples. The threshold cycle (C
T
)
was automatically determined for each reaction by the system
set with default parameters. The specificity of the PCR was
determined by melt curve analysis of the amplified products
using the standard method installed in the system.
Treatments with growth hormones and abiotic stresses
Two-week-old plants grown on MS-agar plates were trans-
ferred to MS liquid cultures supplemented with various
growth hormones, including methyl jasmonate (mJA) or 1-
aminocyclopropane-1-carboxylic acid (ACC) (20 lM each,
unless otherwise specified), for the indicated time periods,
and plant materials were harvested for total RNA extrac-
tion. ABA was used at a final concentration of either 1 or
5 lM for MS-agar plates or 20 lM for MS liquid cultures.
For the assays on the effects of drought on gene expres-
sion, 2-wk-old plants grown on MS-agar plates were put on
a dry 3MM paper and incubated at room temperature for
the indicated time periods. For the assays on the effects of
high salinity on gene expression, 2-wk-old plants grown on
MS-agar plates were soaked in MS liquid cultures contain-
ing 200 mM NaCl and incubated with gentle shaking
under constant light for the indicated time periods.
Assays on pathogen infection
Bacterial cells of P. syringae pv. tomato strain DC3000 were
cultured for 2 d at 28C in King’s B medium supplemented
with rifampicin (50 lg l
)1
) (Park et al., 2007). A bacterial
cell suspension was prepared at 10
7
cfu ml
)1
in 10 mM
MgCl
2
supplemented with 250 ppm TWEEN 80 and
sprayed directly on to the leaf surface. After incubation for
16 h at 25C and 100% relative humidity in complete
darkness, the inoculated plants were transferred to a growth
chamber set at 23C and 80% relative humidity and grown
further under long days. Measurements of bacterial cell
growth were carried out as previously described (Park et al.,
2007) using whole leaves of 4-wk-old plants grown in soil.
For the direct infiltration assays, bacterial cells of P. syrin-
gae pv. tomato strain DC3000 were prepared as described
(Park et al., 2007). Bacterial cells were collected and resus-
pended in resuspension buffer containing 10 mM MgCl
2
.
The sixth leaves of 4-wk-old plants grown in soil were infil-
trated with the bacterial cell suspensions by injecting into
the abaxial side of the leaves using 1 ml needleless syringes.
Analysis of anthocyanin concentrations
Extraction and quantification of anthocyanins from the leaf
tissues were carried out as described previously (Rabino &
Mancinelli, 1986). Plant leaves were homogenized in liquid
nitrogen, and anthocyanins were extracted using methanol
that contained 1% HCl (v ⁄ v). Extraction steps were con-
ducted at 4C. After centrifugation, the supernatant was
used for measurements of absorbance at 530 and 657 nm.
The formula A
530
– 0.25 · A
657
was used to calculate the
amounts of anthocyanins.
Measurements of endogenous SA contents
Extraction and quantification of endogenous SA and SAG
from the leaf tissues of 2-wk-old plants grown on MS-agar
plates were carried out as described previously (Bowling
et al., 1994). Three independent measurements were aver-
aged. Statistical significance was determined using Student’s
t-test.
Results
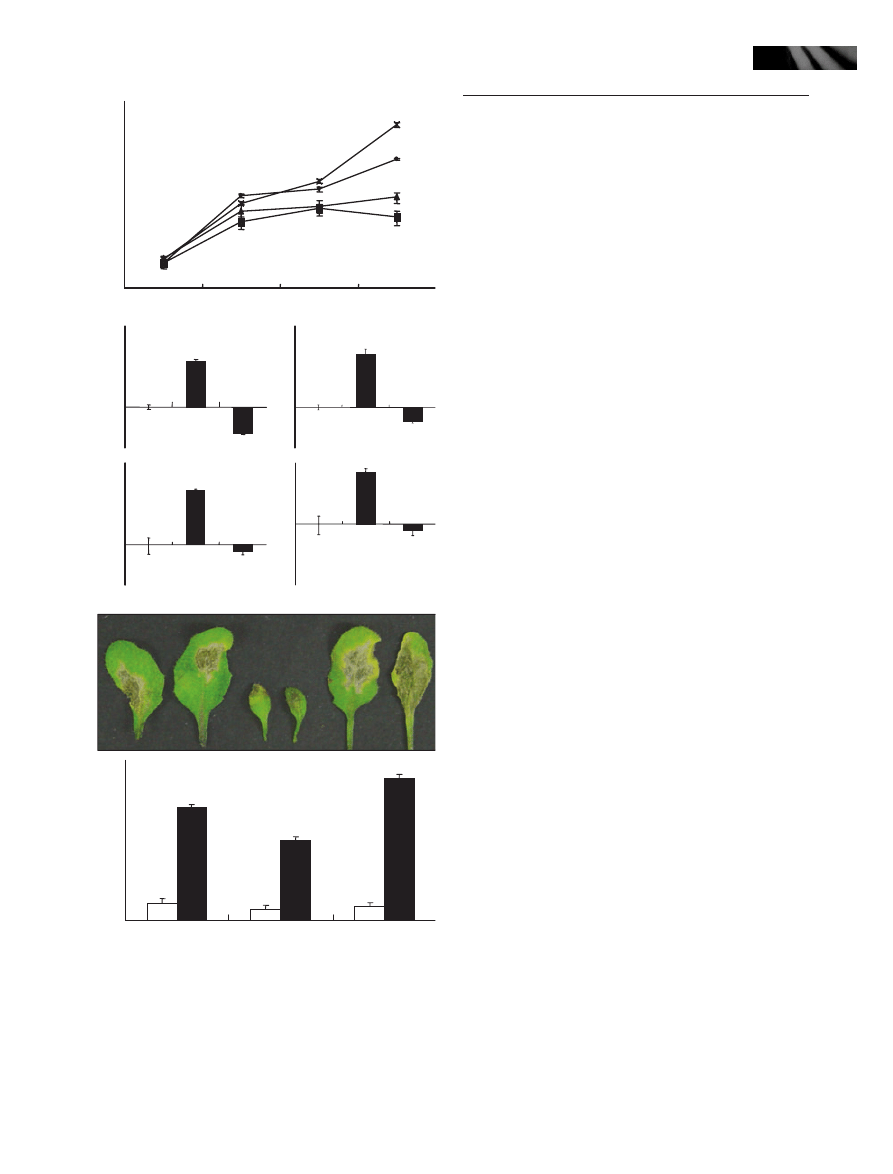
Anthocyanins accumulate in the myb96-1d leaves
We have recently reported that the MYB96 transcription
factor is intimately related with ABA-mediated drought
New
Phytologist
Research
473
The Authors (2010)
Journal compilation New Phytologist Trust (2010)
New Phytologist (2010) 186: 471–483
www.newphytologist.com

stress responses, particularly during lateral root develop-
ment (Seo et al., 2009). The activation-tagging myb96-1d
line is characterized by having reduced growth with dis-
turbed leaf morphology and by exhibiting an enhanced
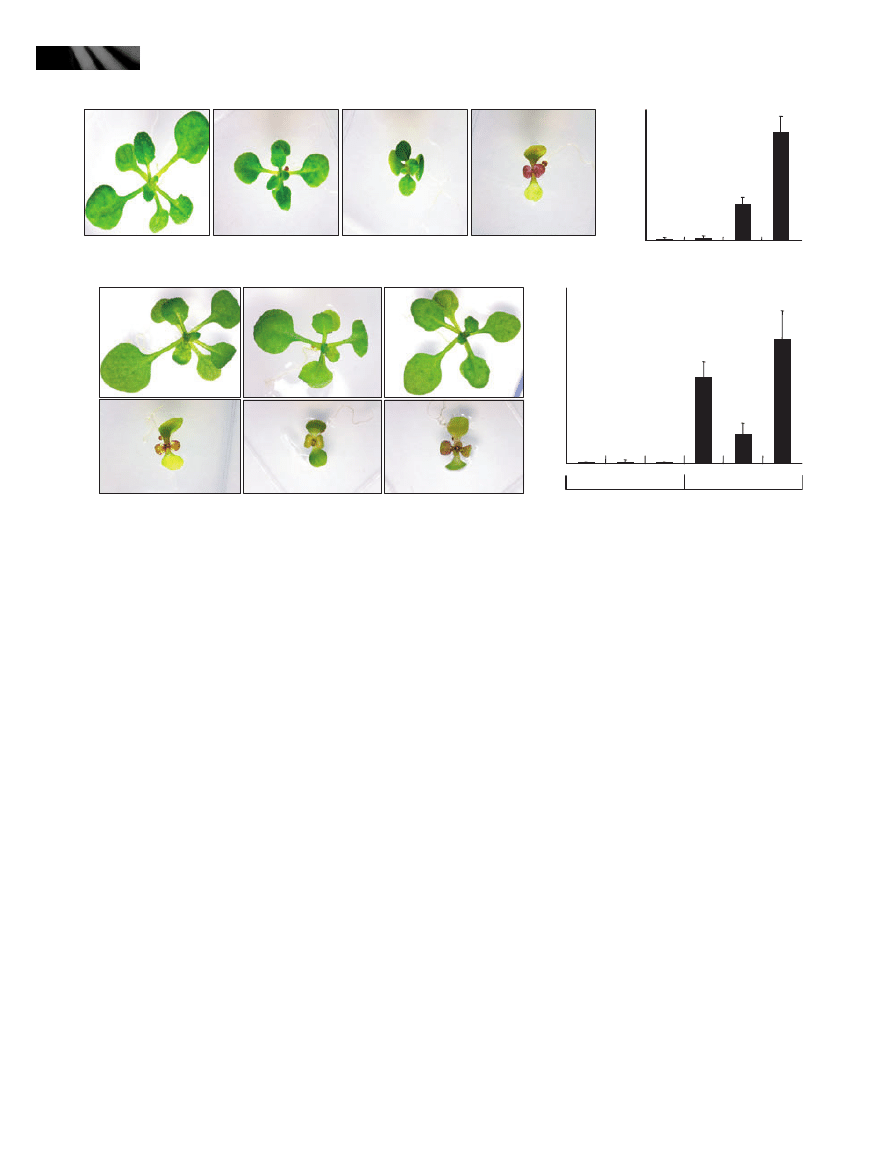
resistance to drought (Fig. 1a; Seo et al., 2009). Notably,
we also found that anthocyanins accumulate to a high
concentration in the leaves of older myb96-1d plants
(Fig. 1a, right panels). There were no discernible differ-
ences in the amounts of anthocyanin during the seedling
growth stage. However, it was significantly elevated in
the later growth stages, evidently 32 d after germination.
Anthocyanin accumulation was initiated in the leaf mar-
gin of the myb96-1d line and later spread throughout
the whole leaf area (Fig. 1a, right panels). Anthocyanin
content was higher by c. 80-fold in the myb96-1d leaves
than in wild-type leaves (Fig. 1b). By contrast, chloro-
phyll content was not discernibly changed in the leaves
of the activation-tagging myb96-1d line (Fig. 1c). The
MYB96-deficient mutant, myb96-1, had indistinguishable
phenotypes compared with wild-type plants under nor-
mal growth conditions (Seo et al., 2009).
We examined expression patterns of the genes encoding
anthocyanin biosynthetic enzymes and constituting the
MBW complex by qRT-PCR. We found that the PAP1
and PAP2 genes were significantly up-regulated in the acti-
vation-tagging myb96-1d line (Fig. 1d; Fig. S1). The SA
biosynthetic enzyme gene PAL1 and the DIHYDROFL-
AVONOL 4-REDUCTASE (DFR) gene that catalyzes the
conversion of dihydroquercetin to leucocyanidin in the
anthocyanin biosynthesis were also induced by c. two- to
threefold in the activation-tagging myb96-1d plant. These
observations indicate that the MYB96-mediated signaling
induces a subset of anthocyanin biosynthetic genes, result-
ing in a high accumulation of anthocyanins in the myb96-
1d plants.
Col-0
(a)
(b)
(d)
(c)
myb96-1d
(+/–)
myb96-1d
(+/+)
35S:
MYB96
0
20
40
60
80
100
120
140
Rel. chlorophyll content (%)
0
20
40
60
80
100
120
140
Col-0
Rel. anthocyanin content
PAP1
0
1
2
3
4
5
6
7
8
9
PAL1
0
1
2
3
PAP2
0
1
2
3
4
5
6
7
8
DFR
0
1
2
3
4
myb96-1d myb96-1
Col-0 myb96-1d myb96-1
Col-0 myb96-1d myb96-1
Col-0 myb96-1d myb96-1
Col-0 myb96-1d myb96-1
Col-0 myb96-1d myb96-1
Fold change relative
to
eIF4a
Fold change relative
to
eIF4a
*
*
*
*
*
Fig. 1 Anthocyanins accumulate to a high concentration in the
leaves of the MYB96-overexpressing myb96-1d activation-tagging
line. (a) High accumulation of anthocyanins in the adult leaves of
the activation-tagging myb96-1d line and 35S:MYB96 transgenic
Arabidopsis plants. Five-week-old plants grown in soil were photo-
graphed (left panel). Anthocyanin accumulation is evident, particu-
larly in the rosette leaves of older mutant plants (right, upper panel).
Two representative rosette leaves with visible anthocyanin accumu-
lation are shown (right, bottom panel). Bar, 1 cm. (b) Measurement
of anthocyanins in the mutant leaves. The normalized value of wild-
type plants was used as a reference value of (1) for the relative
anthocyanin contents. (c) Measurements of chlorophyll in the
mutant leaves. Total chlorophyll levels were measured. The normal-
ized value of wild-type plants was used as a reference (100%) for
the comparison of relative chlorophyll contents. In (b) and (c), the
leaves of 5-wk-old plants grown in soil were used for extraction of
anthocyanins and chlorophylls, and five measurements were aver-
aged and statistically treated using Student’s t-test (*P < 0.01). Bars
indicate standard error of the mean. (d) Transcript abundances of
several anthocyanin biosynthetic enzyme genes in the mutants
(PAL1, DFR, PAP1 and PAP2). Whole plants grown on MS-agar
plates for 2 wk were used for extraction of total RNAs. Transcript
abundances were compared by quantitative real-time RT-PCR (qRT-
PCR). Biological triplicates and technical duplicates were averaged
and statistically treated (t-test, *P < 0.01). Bars indicate standard
error of the mean. A MYB96-deficient mutant myb96-1 was also
included in the assays of (b)–(d).
474
Research
New
Phytologist
The Authors (2010)
Journal compilation New Phytologist Trust (2010)
New Phytologist (2010) 186: 471–483
www.newphytologist.com
The activation-tagging myb96-1d line and the
myb96-1 mutant exhibit altered resistance responses
to pathogen infection
Anthocyanin accumulation in plant tissues is closely related
to abiotic and biotic stress responses (Dixon & Paiva, 1995;
Chalker-Scott, 1999). It is sometimes used as a physiologi-
cal marker for disease resistance response in some plant spe-
cies (He & Dixon, 2000; Dixon, 2001). In addition, we
found that a SA biosynthetic enzyme gene, PAL1, is up-reg-
ulated in the activation-tagging myb96-1d line. Based on
previous observations as well as our own, it was hypothe-
sized that the MYB96 gene might be related to pathogen
resistance response in addition to its role in drought resis-
tance.
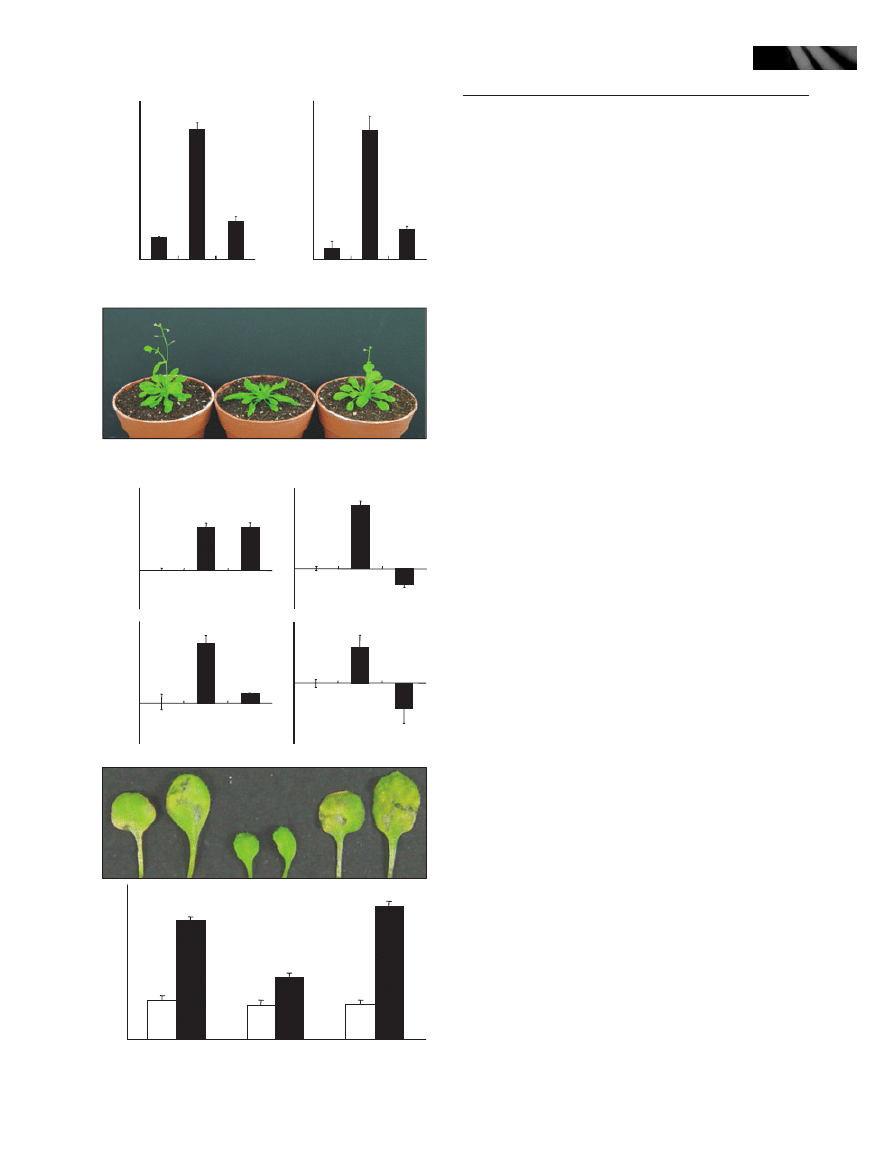
To examine the hypothesis, we infected the myb96-1d
and myb96-1 plants with a virulent P. syringae DC3000
strain. As expected, counting of bacterial cell growth in the
infected mutants revealed that, whereas the MYB96-overex-
pressing myb96-1d line exhibited an enhanced disease resis-
tance to pathogen infection, the myb96-1 mutant was
relatively more susceptible to pathogen infection (Fig. 2a).
Transgenic plants overexpressing the MYB96 gene under
the control of the Cauliflower Mosaic Virus (CaMV) 35S
promoter were also resistant to pathogen infection similar
to the activation-tagging myb96-1d line.
Consistent with the altered resistance responses to patho-
gen infection in the mutants, expression of several PR genes
were changed in the mutants. We observed that while the
transcript abundances of PR1, PR2, and PR5, which partici-
pate in SA signaling (Seo et al., 2008), were significantly
higher in the activation-tagging myb96-1d line, they were
slightly but reproducibly lower in the myb96-1 mutant
(Fig. 2b).
We have recently reported that MYB96 regulates drought
resistance by modulating stomatal opening. It has been
known that stomatal closure is triggered in response to bac-
terial infection and pathogen-associated molecular pattern
(PAMP), which serves as part of the plant defense mecha-
nisms to restrict bacterial invasion (Melotto et al., 2006). It
4
5
6
7
8
9
10
(a)
(b)
(c)
0
2
4
6(dpi)
Log (cfu per 0.1 g FW)
*
PR1
0.1
1
10
100
Col-0
myb96-1d
myb96-1
MYB96
0.1
1
10
100
Col-0
myb96-1d
myb96-1
PR2
0.1
1
10
100
Col-0
myb96-1d
myb96-1
*
*
*
*
*
*
*
0.1
1
10
PR5
Col-0
myb96-1d
myb96-1
*
4
5
6
7
8
9
10
Log (cfu per 0.1 g FW)
Col-0
myb96-1d
myb96-1
Col-0
myb96-1d
myb96-1
*
*
Fold change/
eIF4a
Fold change/
eIF4a
Fig. 2 The activation-tagging myb96-1d line exhibits an enhanced
resistance to Pseudomonas syringae infection. (a) Pathogen infec-
tion assays on the myb96-1d and myb96-1 plants. Arabidopsis
plants were infected with a virulent P. syringae strain by spray inocu-
lation, and the numbers of bacterial cells were counted. Four-week-
old plants grown in soil were used for infection assays. Five indepen-
dent countings were averaged and statistically treated (t-test,
*P < 0.01). Bars indicate standard error of the mean. (b) Expression
of the pathogenesis-related (PR) genes in the activation-tagging
myb96-1d line and the myb96-1 mutant. Whole plants grown on
MS-agar plates for 2 wk were used for extraction of total RNAs.
Transcript abundances were compared by quantitative real-time RT-
PCR (qRT-PCR). Biological triplicates and technical duplicates were
averaged and statistically treated (t-test, *P < 0.01). Bars indicate
standard error of the mean. The y-axis was presented on a logarith-
mic scale for better comparison of fold changes. (c) Pathogen infec-
tion assays on the myb96-1d and myb96-1 leaves. The sixth leaves
of 4-wk-old plants grown in soil were used for infiltration assays.
Three independent experiments were averaged and statistically
treated (t-test, *P < 0.01). Bars indicate standard error of the mean.
Open bars, 0 d postinfection (dpi); closed bars, 4 dpi.
New
Phytologist
Research
475
The Authors (2010)
Journal compilation New Phytologist Trust (2010)
New Phytologist (2010) 186: 471–483
www.newphytologist.com

was therefore suspected that the reduced stomatal aperture
in the activation-tagging myb96-1d line would be related to
the observed disease resistance. To answer the question, we
infiltrated P. syringae pv. tomato DC3000 directly into the
leaves of the myb96-1d and myb96-1 plants. While the acti-
vation-tagging myb96-1d line exhibited an enhanced resis-
tance, the myb96-1 mutant showed a higher susceptibility
(Fig. 2c), indicating that the reduced or increased stomatal
closure does not contribute significantly to the altered resis-
tance responses in the myb96-1d and myb96-1 leaves.
Together, these observations indicate that the MYB96 gene
is intimately related with disease resistance response in
plants.
The SID2 gene is induced in the activation-tagging
myb96-1d line
To investigate the molecular cause underlying the altered
disease resistance responses of the activation-tagging
myb96-1d line and the myb96-1 mutant, we examined the
expression patterns of SA biosynthetic and signaling genes.
The PAL1 gene was induced moderately (2.3-fold) in the
activation-tagging myb96-1d line (Fig. 1d). In particular,
the SID2 gene was significantly up-regulated in the activa-
tion-tagging myb96-1d line but slightly suppressed in the
myb96-1 mutant (Fig. 3a). By contrast, the expression
patterns of other SA biosynthetic genes, such as PAL2,
PAL3, and SID1 ⁄ ICS2, and of SA signaling genes, inclu-
ding GST6, NPR1, and TGA2, were unaffected to a
noticeable degree by the myb96 mutations (Fig. S2). These
observations support the proposition that the MYB96
gene positively regulates the SID2 gene, which would
result in SA accumulation in the activation-tagging
myb96-1d line. The MYB96 gene was uninfluenced by
exogenous SA application (Fig. 3b), indicating that the
MYB96 gene acts upstream of SID2 expression and thus
SA biosynthesis.
Our data suggest that the MYB96 gene might be
induced by pathogen infection and the MYB96 induction
would occur earlier than induction of SA biosynthesis
and PR gene induction. To examine this possibility, we
investigated expression kinetics of the MYB96,
SID2,
and PR1 genes after treatment with the bacterial flagellin
peptide elicitor (Flg22), which efficiently elicits plant
defense responses (Takai et al., 2008). The MYB96 gene
was rapidly induced by the Flg22 treatment, and the
transcript abundance reached the peak within 1 h
(Fig. 3c). The SID2 induction was initiated 2 h after
treatments and reached the peak 6 h after Flg22 applica-
tion. The PR1 induction was initiated 12 h after treat-
ments, supporting the notion that the MYB96 induction
is an early event in plant disease resistance response,
occurring ahead of the promotion of SA biosynthesis and
PR gene induction.
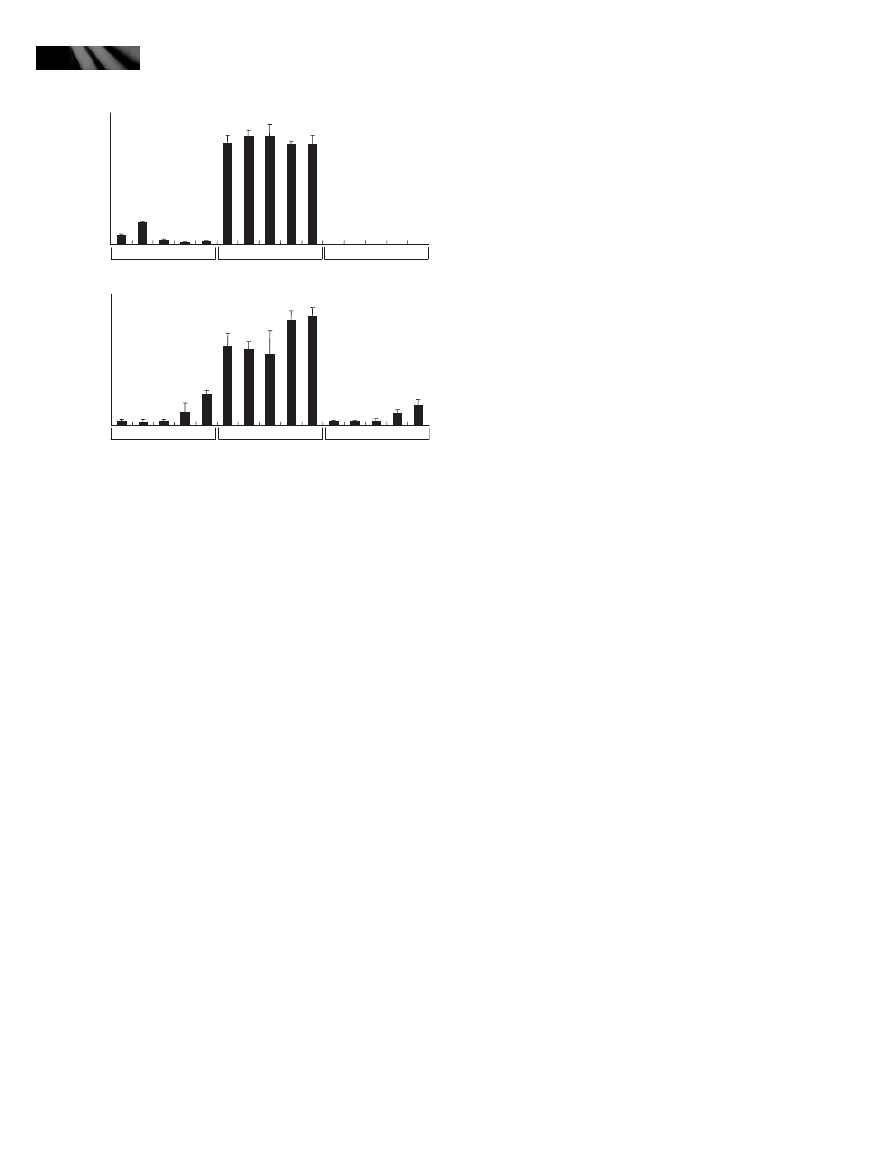
Endogenous concentrations of SA and SAG are
elevated in the activation-tagging myb96-1d line
Enhanced disease resistance and up-regulation of the SA
biosynthetic enzyme gene SID2 in the activation-tagging
myb96-1d line suggested that SA biosynthesis would be ele-
vated in the mutant. We therefore measured the endoge-
nous concentrations of SA and SAG in the myb96-1d and
myb96-1 plants. As expected, the endogenous concentra-
tions of free SA and conjugated SA (SAG) were c. sevenfold
and 10-fold higher, respectively, in the activation-tagging
myb96-1d line (Fig. 4a). The concentrations of SA and
SAG in the myb96-1 mutant were not detectably different
from those in wild-type plants. This might be the result of
functional redundancy among the multiple MYB transcrip-
tion factors functioning in pathogen resistance response.
SID2
0
2
4
6
8
10
12
(a)
(c)
(b)
Col-0
0
0.2
0.4
0.6
0.8
1
1.2
1.4
1.6
MYB96
myb96-1d myb96-1
Mo2
Mo6
SA2
SA6
0.1
1
10
100
0
1
2
6
12
24
MYB96
SID2
PR1
Time after flg22 treatment (h)
*
Fold change relative
to
eIF4a
Fold change relative to
eIF4a
Fig. 3 The SALICYLIC ACID INDUCTION DEFICIENT2 (SID2) gene
is up-regulated in the activation-tagging myb96-1d line. Two-week-
old, whole Arabidopsis plants grown on MS-agar plates were used
for extraction of total RNAs or for subsequent treatment with sali-
cylic acid (SA) or flagellin22 (Flg22). Transcript abundances were
compared by quantitative real-time RT-PCR (qRT-PCR). Biological
triplicates and technical duplicates were averaged and statistically
treated (t-test, *P < 0.01). Bars indicate standard error of the mean.
(a) Up-regulation of the SID2 gene in the activation-tagging
myb96-1d line. (b) Effects of SA on MYB96 expression. Plants were
incubated with gentle shaking in liquid MS cultures supplemented
with 0.1 mM SA for the indicted time periods before extraction of
total RNAs. Mo, mock. (c) Expression kinetics of the MYB96, SID2,
and pathogenesis-related1 (PR1) genes after application of 1 lM
Flg22. Peaks of the transcript abundances for individual genes are
indicated by arrows. The relative expression levels were fold changes
relative to the transcript abundances at 0 h time points for each
gene.
476
Research
New
Phytologist
The Authors (2010)
Journal compilation New Phytologist Trust (2010)
New Phytologist (2010) 186: 471–483
www.newphytologist.com
Our data indicate that the increased SA biosynthesis con-
tributes to the myb96-1d phenotypes, such as enhanced dis-
ease resistance and altered leaf morphology. To confirm the
intimate relationship between SA and phenotypic altera-
tions in the activation-tagging myb96-1d line, we genetically
crossed the mutant with SA-deficient NahG transgenic
plants that express a salicylate hydroxylase (Friedrich et al.,
1995). Strikingly, the myb96-1d phenotypes, such as
impaired leaf morphology and growth retardation, disap-
peared in the myb96-1d X NahG cross (Fig. 4b) as well as
in the myb96-1d X sid2 cross (Fig. S3). In addition, the
transcript abundances of the PR1, PR2, and PR5 genes were
reduced in the myb96-1d X NahG cross to an amount com-
parable to or even lower than those observed in wild-type
plants (Col-0) (Fig. 4c). By contrast, the transcript abun-
dance of the MYB96 gene was unaffected in the genetic
cross, which is consistent with the lack of SA effects on the
MYB96 expression (Fig. 3b).
To further confirm the role of MYB96 in disease resis-
tance, we infected the myb96-1d X Col-0 and myb96-1d X
NahG crosses with P. syringae pv. tomato DC3000 cells by
spray inoculation. Consistent with the expression patterns
of the PR genes (Fig. 4c), whereas the myb96-1d X Col-0
cross showed an enhanced resistance, the disease resistance
of the myb96-1d X NahG cross was compromised to a
degree comparable to that observed in wild-type plants
(Fig. 4d). It is therefore evident that the myb96-1d pheno-
types are, at least in part, attributable to elevated SA biosyn-
thesis in the mutant.
The MYB96-mediated ABA signals confer an enhanced
drought resistance via a RD22-mediated pathway (Seo
Col-0
0
200
400
600
800
1000
1200
1400
1600
(a)
(b)
(c)
(d)
ng g
–1
FW
ng g
–1
FW
Free SA
Col-0
myb96-1dmyb96-1
Col-0
myb96-1dmyb96-1
0
2000
4000
6000
8000
10 000
12 000
SAG
*
*
0.1
1
10
100
1
10
100
PR1
0.1
1
10
100
Col-0
PR5
0.1
1
10
Col-0
PR2
Col-0
MYB96
Col-0
*
*
*
*
*
*
*
4
5
6
7
8
9
Log (cfu per 0.1 g FW)
Col-0
myb96-1d X Col-0
myb96-1d X Col-0
myb96-1d
X Col-0
myb96-1d
X Col-0
myb96-1d
X Col-0
myb96-1d
X NahG
myb96-1d
X Col-0
myb96-1d X Col-0
(F1)
myb96-1d X NahG
(F1)
myb96-1d
X NahG
myb96-1d
X NahG
myb96-1d
X NahG
myb96-1d X NahG
myb96-1d X NahG
Col-0
*
*
Fold change relative
to
eIF4a
Fold change relative
to
eIF4a
Fig. 4 Endogenous concentrations of salicylic acid (SA) and its con-
jugated form SA-b-glucoside (SAG) are elevated in the activation-
tagging myb96-1d line. (a) Endogenous contents of SA and SAG in
the activation-tagging myb96-1d line and the myb96-1 mutant. The
leaves of 2-wk-old Arabidopsis plants grown on MS-agar plates
were used for extraction of SA and SAG. Three measurements were
averaged and statistically treated (t-test, *P < 0.01). Bars indicate
standard error of the mean. (b) Phenotypic comparison of the
myb96-1d X Col-0 and myb96-1d X NahG genetic crosses. The acti-
vation-tagging myb96-1d line was crossed with the SA-deficient
NahG transgenic plants. Four-week-old plants grown in soil were
photographed. (c) Transcript abundances of the pathogenesis-
related (PR) genes in the genetic crosses. Two-week-old plants
grown on MS-agar plates were used for extraction of total RNAs.
Transcript abundances were compared by quantitative real-time RT-
PCR (qRT-PCR). Biological triplicates and technical duplicates were
averaged and statistically treated (t-test, *P < 0.01). Bars indicate
standard error of the mean. The y-axis was presented on a logarith-
mic scale for better comparison of fold changes. (d) Pathogen infec-
tion assays on the myb96-1d X Col-0 and myb96-1d X NahG
genetic crosses. Four-week-old plants grown in soil were used for
the spray inoculation assays. The third and fourth leaves were pho-
tographed (upper panel). Three independent experiments were
averaged and statistically treated (t-test, *P < 0.01). Bars indicate
standard error of the mean (bottom panel). Open bars, 0 d postin-
fection (dpi); closed bars, 5 dpi.
New
Phytologist
Research
477
The Authors (2010)
Journal compilation New Phytologist Trust (2010)
New Phytologist (2010) 186: 471–483
www.newphytologist.com
et al., 2009). However, expression of the RD22 gene was
unaltered in the myb96-1d X NahG cross (Fig. S4), indicat-
ing that the MYB96-mediated regulation of the RD22 gene
during drought resistance response is functionally separated
from the MYB96-mediated regulation of SA biosynthesis
and thus of pathogen resistance response.
MYB96 plays a role in pathogen induction of PR genes
The activation-tagging myb96-1d line showed an enhanced
pathogen resistance, and a small group of PR genes are up-
regulated in the mutant. The concentration of endogenous
SA content was also elevated in the activation-tagging
myb96-1d line. A critical question was therefore whether
the MYB96 gene contributes to the PR induction in
infected plants.
To examine this, wild-type, myb96-1d, and myb96-1
plants were infected with P. syringae cells by spray inocula-
tion, and the PR1 expression kinetics were investigated by
qRT-PCR after infection. The MYB96 gene was rapidly
induced after pathogen infection, and the transcript abun-
dance decreased gradually after the peak at 12 h (Fig. 5a).
Under the same infection conditions, the PR1 gene induc-
tion was initiated 48 h after infection, and the transcript
abundance was further elevated throughput the time course.
The PR1 gene was induced in a similar kinetics in the
infected myb96-1 mutant. However, the PR1 transcript
abundance was lower by c. 40% in the myb96-1 mutant
than in wild-type plants (Fig. 5b). It is well known that the
PR1 gene is induced after pathogen infection in a SA-
dependent manner (Bol et al., 1990). It was therefore con-
cluded that at least a portion of the PR1 induction after
pathogen infection depends on a functional MYB96 activ-
ity.
MYB96-mediated abiotic stress signals induce the
SID2 gene in an ABA-dependent manner
The MYB96 gene is a component of ABA signaling in
drought stress response. Therefore, related questions were
whether ABA and abiotic stresses affect the SID2 expression
and, if so, whether the effects of abiotic stresses on the SID2
expression depend on ABA.
Wild-type plants were treated with ABA and mannitol
that confers osmotic stress on plants, and the transcript
abundances of the SID2 gene were examined by qRT-PCR.
The SID2 expression was induced by more than fourfold in
the presence of ABA (Fig. 6a, left panel). Mannitol treat-
ment also exhibited a similar effect on the SID2 expression
(Fig. 6a, right panel). However, the inductive effect of ABA
on the SID2 expression was reduced by more than 30% in
the myb96-1 mutant (Fig. 6b), indicating that a functional
MYB96 activity is required for the ABA induction of the
SID2 gene.
Drought and high salinity also showed inductive effects
on the SID2 expression (Fig. 6c). In addition, the effects of
high salinity on the SID2 expression were reduced by c.
40% in the ABA-deficient aba3-1 mutant (Fig. 6d), indi-
cating that abiotic stresses induce the SID2 gene, at least in
part, via the ABA-dependent pathway. Together, our obser-
vations indicate that ABA-mediated abiotic stress signals
regulate the SID2 gene and that the MYB96 gene plays a
role in these signaling cascades.
MYB96 serves as a molecular link that integrates ABA
and SA signals
We found that the activation-tagging myb96-1d line pheno-
types, such as dwarfed growth with altered leaf morphology,
are efficiently rescued in the myb96-1d X NahG cross
(Fig. 4b). The transcript abundances of the PR genes and
disease resistance were also compromised in the cross. Fur-
thermore, the activation-tagging myb96-1d line exhibits an
enhanced resistance to drought (Seo et al., 2009) as well as
to pathogen infection (Fig. 2a). These observations strongly
support the proposition that the ABA and SA signals are
closely linked, and the myb96-1d phenotypes, such as
anthocyanin accumulation, would be caused by simulta-
neous stimulation of both ABA and SA responses in the
mutant.
PR1
0
5
10
15
20
25
30
35
MYB96
0
2
4
6
8
10
12
14
16
(a)
(b)
Col-0
myb96-1d
0 h 12 h 24 h 48 h 72 h
0 h 12 h 24 h 48 h 72 h
0 h 12 h 24 h 48 h 72 h 0 h 12 h 24 h 48 h 72 h
0 h 12 h 24 h 48 h 72 h 0 h 12 h 24 h 48 h 72 h
myb96-1
Col-0
myb96-1d
myb96-1
Fold change relative to
eIF4a
Fold change relative to
eIF4a
Fig. 5 The MYB96 gene contributes to the pathogenesis-related
(PR) induction by pathogen infection. Four-week-old Arabidopsis
plants grown in soil were infected with a virulent Pseudomonas sy-
ringae strain. Transcript abundances of the MYB96 gene (a) and of
the PR1 gene (b) were compared by quantitative real-time RT-PCR
(qRT-PCR) at the indicated time points after infection. Biological
triplicates and technical duplicates were averaged. Bars indicate
standard error of the mean.
478
Research
New
Phytologist
The Authors (2010)
Journal compilation New Phytologist Trust (2010)
New Phytologist (2010) 186: 471–483
www.newphytologist.com
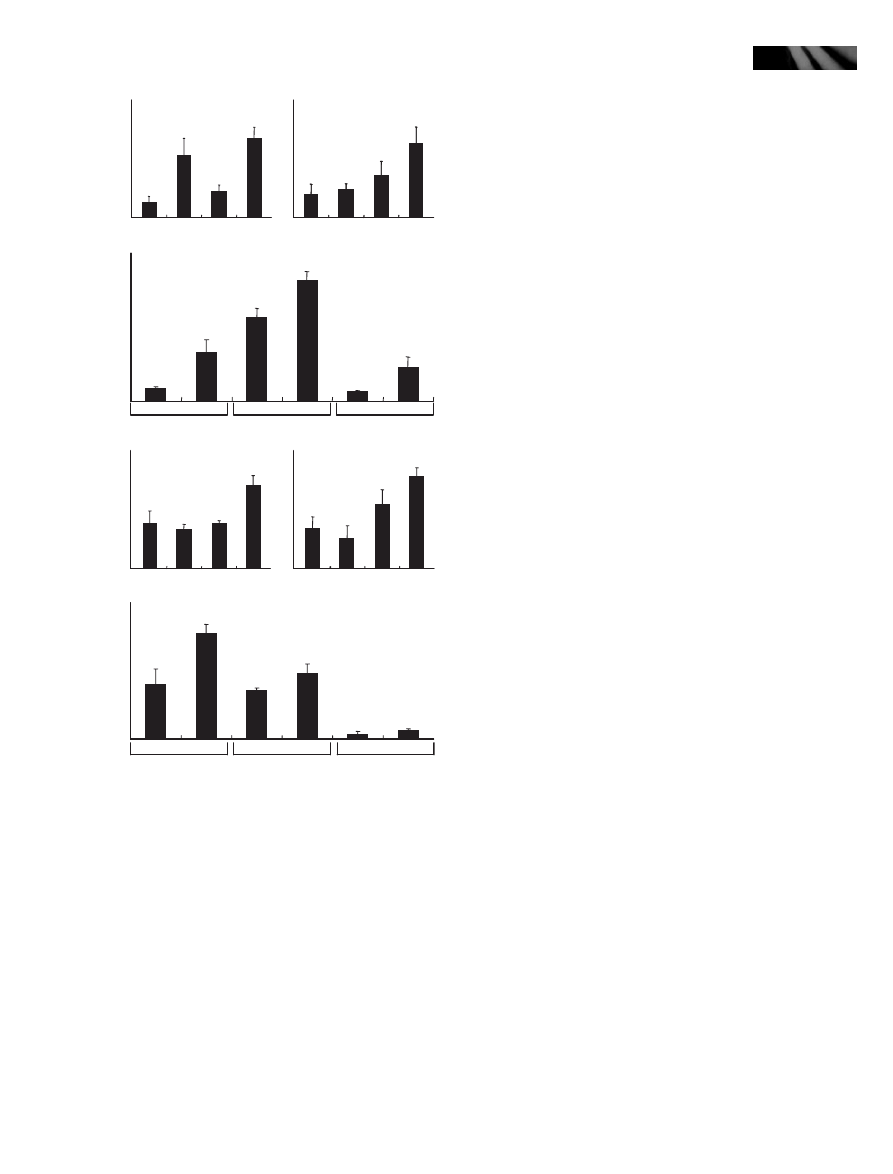
To examine this, wild-type plants were treated with SA
(0.1 mM) or NaCl (150 mM), or both, and plant pheno-
types were analyzed. Plant growth was delayed in the pres-
ence of either SA or NaCl with a more severe retardation in
the NaCl-treated plants (Fig. 7a, left panel). When the
plants were treated with both SA and NaCl, a remarkable
amount of anthocyanins was accumulated in the leaves,
similar to that observed in the myb96-1d leaves (Fig. 7a,
right panel), supporting the notion that the myb96-1d phe-
notypes are caused by an additive effect of ABA and SA sig-
nals.
We next treated an ABA signaling mutant, abi1-1, and
the NahG transgenic plants with both SA and NaCl. Plant
growth was severely delayed in all the treated plants
(Fig. 7b). However, anthocyanins accumulated to a high
concentration only in the NahG transgenic plants, but the
accumulation was greatly reduced in the abi1-1 mutant,
indicating that anthocyanin accumulation is regulated pri-
marily by ABA-mediated signaling (see the following sec-
tion).
Discussion
Two major stress hormones functioning under biotic and
abiotic stress conditions are ABA and SA. Accumulating
evidence demonstrates that the two hormones act either
individually or through intricate signaling crosstalks (Park
et al., 2007; Flors et al., 2008; Yasuda et al., 2008), reflect-
ing that a finely tuned hormone balance is critical for plant
survival under stress conditions.
Abscisic acid is generally considered as a negative regula-
tor of disease resistance. Exogenous application of ABA is
correlated with an increased susceptibility to pathogen
infection, and ABA-deficient mutants exhibit an enhanced
pathogen resistance (Mauch-Mani & Mauch, 2005; Fan
et al., 2009). In another case, while elevated concentrations
of SA are required to build up an innate immune response,
bacterial effectors rapidly activate ABA biosynthesis in
plants to suppress defense responses (de Torres-Zabala
et al., 2007). In this signaling scheme, ABA antagonizes
SA-mediated defense responses, providing a mechanistic
base for priming events during plant defense responses.
Although antagonistic interactions have been reported
between ABA and SA, recent studies imply that positive
interactions between the ABA signaling pathway and the
biotic signaling network involving SA, jasmonic acid (JA)
and ethylene (ET) enhance a tolerance response to abiotic
and biotic stresses. It has been recently proven that plant
pathogens take advantage of ABA signaling pathways to
promote pathogenesis (Mengiste et al., 2003; Chini et al.,
2004). The BOTRYTIS SUSCEPTIBLE1 (BOS1) gene con-
trols both JA- and ABA-inducible genes. As a result, a loss-
of-function bos1 mutant is susceptible to both necrotrophic
pathogens and osmotic and oxidative stresses (Mengiste
et al., 2003). The ADR1 gene is an another intriguing
example supporting an intimate functional relationship
between abiotic and biotic stress responses. While most of
0
1
2
3
0
1
2
3
NaCl2 NaCl6
Mock1
Mock2 Mock6
Mock2 DR1
DR2
0
1
2
3
4
5
6
7
8
(a)
(b)
(c)
(d)
0
1
2
3
4
5
0
2
4
6
8
10
12
Mock
ABA
SA ABA+SA
Mock2 Mock6 Man2 Man6
Mock
ABA
Mock
ABA
Mock
ABA
Col-0
myb96-1d
myb96-1
0
1
2
Col-0
Mock
NaCl
Mock
NaCl
Mock
NaCl
aba3-1
sid2
*
*
*
*
*
*
*
*
*
SID2
SID2
SID2
SID2
SID2
SID2
Fold change
relative to
eIF4a
Fold change
relative to
eIF4a
Fold change
relative to
eIF4a
Fold change
relative to
eIF4a
Fig. 6 The MYB96 gene modulates abscisic acid (ABA)-mediated
abiotic stress signals in inducing the SALICYLIC ACID INDUCTION
DEFICIENT2 (SID2) gene. Two-week-old Arabidopsis plants grown
on MS-agar plates were used for treatments with ABA, salicylic acid
(SA), mannose (Man), NaCl, or drought (DR). Whole plants were
used for extraction of total RNAs. Transcript abundances were com-
pared by quantitative real-time RT-PCR (qRT-PCR). Biological tripli-
cates and technical duplicates were averaged and statistically treated
(t-test, *P < 0.01). Bars indicate standard error of the mean. (a)
Effects of ABA and SA (left panel) and mannitol (right panel) on the
SID2 expression. (b) Effects of ABA on SID2 expression in the
myb96-1 mutant. (c) Effects of drought (left panel) and NaCl (right
panel) on SID2 expression. (d) Effects of NaCl on SID2 expression in
the aba3-1 and sid2 mutants.
New
Phytologist
Research
479
The Authors (2010)
Journal compilation New Phytologist Trust (2010)
New Phytologist (2010) 186: 471–483
www.newphytologist.com
disease-resistant mutants do not exhibit an enhanced resis-
tance to abiotic stresses, such as drought and high salinity
(Chini et al., 2004), the activation-tagging mutant adr1
exhibits an enhanced resistance to both virulent pathogens
and drought stress. We also observed similar phenotypes in
the activation-tagging myb96-1d line. The ABA-mediated
MYB96 regulation of SA biosynthesis might be another
route for balancing plant responses to pathogen infection
and abiotic stress condition.
In this work, we examined pathogen resistance responses
of the activation-tagging myb96-1d line and the myb96-1
mutant. Expression levels of the SA biosynthetic and signal-
ing genes were also examined. We found that the activa-
tion-tagging myb96-1d line, which has previously been
shown to exhibit an enhanced resistance to drought (Seo
et al., 2009), was also resistant to pathogen infection. By
contrast, the T-DNA insertional myb96-1 mutant was sus-
ceptible to drought and pathogen infection. Interestingly,
the SID2 gene was up-regulated, and the concentrations of
endogenous SA were elevated in the activation-tagging
myb96-1d line, indicating that the enhanced pathogen resis-
tance of the mutant is derived from increased SA biosynthe-
sis. ABA and abiotic stress conditions, such as drought,
osmotic stress, and high salinity, also induced the SID2
gene. However, the inductive effects of ABA were reduced
in the myb96-1 mutant, indicating that the MYB96 gene is,
at least in part, required for the SID2 induction by ABA-
mediated abiotic stress signals.
Our observations demonstrate that the MYB96 transcrip-
tion factor acts as a signaling link that integrates ABA and
SA signals and regulates a synergistic interaction between
the two stress hormones. This scheme is also consistent with
the improved disease resistance of plants exposed to abiotic
stress conditions in Arabidopsis (Gaudet et al., 2003; Grif-
fith & Yaish, 2004). The previous reports (Seo et al., 2009)
and our data indicate that the MYB96 transcription factor
plays diverse roles in plant responses to biotic and abiotic
stresses. It regulates lateral root development under drought
via the ABA-auxin crosstalk and shoot growth and disease
resistance via the ABA–SA interaction. The ABA–SA inter-
action is particularly interesting, because ABA-mediated
abiotic stress signals regulate SA biosynthesis by inducing a
SA biosynthetic enzyme gene, SID2. It will be interesting
whether the ABA–auxin and ABA–SA interactions are
mutually independent or functionally interrelated. Pheno-
typic and molecular analysis of a series of higher-order
mutants would provide insights into how the MYB96 tran-
scription factor modulates the hormonal interactions.
Mock
(a)
(b)
SA 0.1 mM
NaCl 150 mM
SA 0.1 mM
NaCl 150 mM
0
20
40
60
80
100
Rel. anthocyanin content
Mock
SA
NaCl
SA
NaCl
Col-0
abi1-1
NahG
Mock
SA + NaCl
0
20
40
60
80
100
120
140
Col-0
abi1-1
NahG
Col-0
abi1-1
NahG
Rel. anthocyanin content
Mock
SA + NaCl
Fig. 7 The MYB96 gene integrates abscisic acid (ABA) and salicylic acid (SA) signals during plant stress responses. (a) Anthocyanin accumula-
tion in wild-type Arabidopsis plants treated with SA and NaCl. In the presence of SA and NaCl, plant growth is significantly reduced (left panel),
and anthocyanins accumulate to a high concentration (right panel). (b) Anthocyanin accumulation in the abi1-1 mutant and NahG transgenic
plants treated with SA and NaCl. Whereas plant growth is reduced to a similar degree in both plants (left panel), anthocyanin accumulation is
reduced only in the abi1-1 mutant (right panel).
480
Research
New
Phytologist
The Authors (2010)
Journal compilation New Phytologist Trust (2010)
New Phytologist (2010) 186: 471–483
www.newphytologist.com

Abiotic stress-mediated pathogenesis has been widely
documented (Gaudet et al., 2003; Griffith & Yaish, 2004;
Agarwal et al., 2006). A subset of PR genes (PR1, PR2, and
PR5) is also induced by cold, high salt, and drought (Seo
et al., 2008). The PR gene induction is correlated with
enhanced disease resistance in many cases. We also found
that the PR genes were up-regulated, and disease resistance
is improved in the activation-tagging myb96-1d line. How-
ever, the data should be carefully interpreted, and more
works are required to confirm the ABA–SA interaction. It
has been reported that the PR3 gene plays a role in regulat-
ing seed germination in the presence of high salt (Seo et al.,
2008). Other PR genes have also been implicated in various
plant developmental processes (Doxey et al., 2007; Brinin-
stool et al., 2008). It is therefore possible that the PR genes
induced in the activation-tagging myb96-1d line may be
related to a certain developmental process under abiotic
stress conditions, and the altered disease resistance responses
of the myb96-1d and myb96-1 plants would be an indirect
effect.
Moreover, the activation-tagging myb96-1d line exhib-
ited an array of phenotypic alterations, such as delayed
growth and smaller leaves with an altered morphology.
The phenotypic alterations may affect the pathogen resis-
tance response, as previously reported (Calo et al., 2006;
Tang et al., 2007). Cuticular lipids on the leaves, includ-
ing cutin monomers and cuticular waxes, may be chan-
ged in the activation-tagging myb96-1d line. Delayed
growth may also affect the defense responses. Bacterial
cell infiltration assays on the myb96-1d and myb96-1
leaves showed that disturbed leaf morphology and struc-
ture, such as altered stomatal aperature, do not signifi-
cantly affect the resistance responses (Fig. 2c). However,
some doubt still remains, and further studies are required
to resolve the issue.
Additional evidence supporting the role of the MYB96
gene in ABA–SA interaction was provided by the high accu-
mulation of anthocyanin in the activation-tagging myb96-
1d line and in wild-type plants grown in the presence of SA
and NaCl. Anthocyanins accumulate in plants exposed to
diverse biotic and abiotic stress conditions (Winkel-Shirley,
2001, 2002). We observed a high accumulation of anthocy-
anins in the activation-tagging myb96-1d line that exhibits
enhanced resistance responses to both drought and patho-
gen infection (Seo et al., 2009; this work). Plant growth
was severely delayed in the presence of either SA or NaCl.
When wild-type plants were treated with SA and NaCl, an-
thocyanins accumulate to a high concentration in addition
to growth retardation, indicating that anthocyanin accumu-
lation requires both ABA and SA signals. Alternatively, the
ABA and SA signals governing anthocyanin accumulation
might be interconnected.
A notable observation was that while plant growth was
delayed to a similar degree in both the abi1-1 mutant and
the NahG transgenic plants in the presence of high salt and
SA, anthocyanin accumulation was significantly reduced
only in the abi1-1 mutant (Fig. 7b). By contrast, anthocya-
nins still accumulated to a high concentration in the NahG
transgenic plants. This may be the result of the high accu-
mulation of catechol in the NahG transgenic plants. Never-
theless, it is evident that ABA-mediated abiotic stress
signaling plays a primary role in inducing anthocyanin
accumulation.
Acknowledgements
We thank Jae-Yong Ryu for growing plants. This work was
supported
by
the
Brain
Korea
21,
Biogreen
21
(20080401034001), and National Research Laboratory
programs and by grants from the Plant Signaling Network
Research Center (2009-0079297), the Korea Science and
Engineering Foundation (2007-03415), and from the Agri-
cultural R&D Promotion Center (309017-5), Korea Minis-
try for Food, Agriculture, Forestry and Fisheries.
References
Agarwal PK, Agarwal P, Reddy MK, Sopory SK. 2006. Role of DREB
transcription factors in abiotic and biotic stress tolerance in plants. Plant
Cell Reports 25: 1263–1274.
Albrecht V, Weinl S, Blazevic D, D’Angelo C, Batistic O, Kolukisaoglu
U, Bock R, Schulz B, Harter K, Kudla J. 2003. The calcium sensor
CBL1 integrates plant responses to abiotic stresses. Plant Journal 36:
457–470.
Alonso-Ramı´rez A, Rodrı´guez D, Reyes D, Jime´nez JA, Nicola´s G,
Lo´pez-Climent M, Go´mez-Cadenas A, Nicola´s C. 2009. Evidence for a
role of gibberellins in salicylic acid-modulated early plant responses to
abiotic stress in Arabidopsis seeds. Plant Physiology 150: 1335–1344.
Bol JF, Linthorst HJM, Cornelissen BJC. 1990. Plant pathogenesis-
related proteins induced by viral infection. Annual Review of Phyto-
pathology 28: 113–138.
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. 2000. Activation tag-
ging identifies a conserved MYB regulator of phenylpropanoid biosyn-
thesis. Plant Cell 12: 2383–2394.
Borsani O, Valpuesta V, Botella MA. 2001. Evidence for a role of salicylic
acid in the oxidative damage generated by NaCl and osmotic stress in
Arabidopsis seedlings. Plant Physiology 126: 1024–1030.
Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. 1994. A
mutation in Arabidopsis that leads to constitutive expression of systemic
acquired resistance. Plant Cell 6: 1845–1857.
Brininstool G, Kasili R, Simmons LA, Kirik V, Hu¨lskamp M, Larkin JC.
2008. Constitutive expressor of pathogenesis-related Genes5 affects cell
wall biogenesis and trichome development. BMC Plant Biology 8: 58.
Broun P. 2005. Transcriptional control of flavonoid biosynthesis: a com-
plex network of conserved regulators involved in multiple aspects of dif-
ferentiation in Arabidopsis. Current Opinion in Plant Biology 8: 272–279.
Calo L, Garcı´a I, Gotor C, Romero LC. 2006. Leaf hairs influence phyto-
pathogenic fungus infection and confer an increased resistance when
expressing a Trichoderma alpha-1,3-glucanase. Journal of Experimental
Botany 57: 3911–3920.
Chalker-Scott L. 1999. Environmental significance of anthocyanins in
plant stress responses. Photochemistry and Photobiology 70: 1–9.
Chini A, Grant JJ, Seki M, Shinozaki K, Loake GJ. 2004. Drought toler-
ance established by enhanced expression of the CC-NBS-LRR gene,
New
Phytologist
Research
481
The Authors (2010)
Journal compilation New Phytologist Trust (2010)
New Phytologist (2010) 186: 471–483
www.newphytologist.com

ADR1, requires salicylic acid, EDS1 and ABI1. Plant Journal 38: 810–
822.
Denekamp M, Smeekens SC. 2003. Integration of wounding and osmotic
stress signals determines the expression of the AtMYB102 transcription
factor gene. Plant Physiology 132: 1415–1423.
Dixon RA. 2001. Natural products and plant disease resistance. Nature
411: 843–847.
Dixon RA, Paiva NL. 1995. Stress-induced phenylpropanoid metabolism.
Plant Cell 7: 1085–1097.
Doxey AC, Yaish MW, Moffatt BA, Griffith M, McConkey BJ. 2007.
Functional divergence in the Arabidopsis beta-1,3-glucanase gene family
inferred by phylogenetic reconstruction of expression states. Molecular
Biology and Evolution 24: 1045–1055.
Fan J, Hill L, Crooks C, Doerner P, Lamb C. 2009. Abscisic Acid has a
key role in modulating diverse plant-pathogen interactions. Plant
Physiology 150: 1750–1761.
Flors V, Ton J, van Doorn R, Jakab G, Garcı´a-Agustı´n P, Mauch-Mani
B. 2008. Interplay between JA, SA and ABA signalling during basal and
induced resistance against Pseudomonas syringae and Alternaria brassicico-
la. Plant Journal 54: 81–92.
Friedrich L, Vernooij B, Gaffney T, Morse A, Ryals J. 1995. Characteriza-
tion of tobacco plants expressing a bacterial salicylate hydroxylase gene.
Plant Molecular Biology 29: 959–968.
Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward
E, Kessmann H, Ryals J. 1993. Requirement of salicylic acid for the
induction of systemic acquired resistance. Science 261: 754–756.
Garcion C, Lohmann A, Lamodie`re E, Catinot J, Buchala A, Doermann
P, Me´traux JP. 2008. Characterization and biological function of the
ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiology
147: 1279–1287.
Gaudet DA, Laroche A, Frick M, Huel R, Puchalski B. 2003. Plant devel-
opment affects the cold-induced expression of plant defence-related tran-
scripts in winter wheat. Physiological and Molecular Plant Pathology 62:
175–184.
Grant JJ, Chini A, Basu D, Loake GJ. 2003. Targeted activation tagging
of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent
pathogens. Molecular Plant-Microbe Interactions 16: 669–680.
Griffith M, Yaish MW. 2004. Antifreeze proteins in overwintering plants:
a tale of two activities. Trends in Plant Science 9: 399–405.
Gutierrez L, Mauriat M, Gue´nin S, Pelloux J, Lefebvre JF, Louvet R,
Rusterucci C, Moritz T, Guerineau F, Bellini C et al. 2008. The lack
of a systematic validation of reference genes: a serious pitfall undervalued
in reverse transcription-polymerase chain reaction (RT-PCR) analysis in
plants. Plant Biotechnology Journal 6: 609–618.
He XZ, Dixon RA. 2000. Genetic manipulation of isoflavone 7-O-meth-
yltransferase enhances biosynthesis of 4’-O-methylated isoflavonoid
phytoalexins and disease resistance in alfalfa. Plant Cell 12: 1689–
1702.
Ingram J, Bartels D. 1996. The molecular basis of dehydration tolerance
in plants. Annual Review of Plant Physiololgy and Plant Molecular Biology
47: 377–403.
Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi
YD, Cheong JJ. 2008. Overexpression of AtMYB44 enhances stomatal
closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant
Physiology 146: 623–635.
Kim YS, Kim SG, Park JE, Park HY, Lim MH, Chua NH, Park CM.
2006. A membrane-bound NAC transcription factor regulates cell divi-
sion in Arabidopsis. Plant Cell 18: 3132–3144.
Koes R, Verweij W, Quattrocchio F. 2005. Flavonoids: a colorful model
for the regulation and evolution of biochemical pathways. Trends in
Plant Science 10: 236–242.
Lee HI, Leo´n J, Raskin I. 1995. Biosynthesis and metabolism of salicylic
acid. Proceedings of the National Academy of Sciences, USA 92: 4076–
4079.
Lee MM, Schiefelbein J. 1999. WEREWOLF, a MYB-related protein in
Arabidopsis, is a position-dependent regulator of epidermal cell pattern-
ing. Cell 99: 473–483.
Mauch-Mani B, Mauch F. 2005. The role of abscisic acid in plant-patho-
gen interactions. Current Opinion in Plant Biology 8: 409–414.
Melotto M, Underwood W, Koczan J, Nomura K, He SY. 2006. Plant
stomata function in innate immunity against bacterial invasion. Cell
126: 969–980.
Mengiste T, Chen X, Salmeron J, Dietrich R. 2003. The BOTRYTIS
SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor pro-
tein that is required for biotic and abiotic stress responses in Arabidopsis.
Plant Cell 15: 2551–2565.
Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J,
Lee YH, Park CM. 2007. GH3-mediated auxin homeostasis links
growth regulation with stress adaptation response in
Arabidopsis. Journal
of Biological Chemistry 282: 10036–10046.
Rabino I, Mancinelli AL. 1986. Light, temperature, and anthocyanin pro-
duction. Plant Physiology 81: 922–924.
Raffaele S, Rivas S, Roby D. 2006. An essential role for salicylic acid in
AtMYB30-mediated control of the hypersensitive cell death program in
Arabidopsis. FEBS Letters 580: 3498–3504.
Rowan DD, Cao M, Lin-Wang K, Cooney JM, Jensen DJ, Austin PT,
Hunt MB, Norling C, Hellens RP, Schaffer RJ et al. 2009. Environ-
mental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana.
New Phytologist 182: 102–115.
Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville
SC, Manners JM. 2000. Coordinated plant defense responses in Arabid-
opsis revealed by microarray analysis. Proceedings of the National Academy
of Sciences, USA 97: 11655–11660.
Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A,
Nakajima M, Enju A, Sakurai T et al. 2002. Monitoring the expression
profiles of 7000 Arabidopsis genes under drought, cold and high-salinity
stresses using a full-length cDNA microarray. Plant Journal 31: 279–
292.
Seo PJ, Lee AK, Xiang F, Park CM. 2008. Molecular and functional pro-
filing of Arabidopsis pathogenesis-related genes: insights into their roles in
salt response of seed germination. Plant and Cell Physiology 49: 334–
344.
Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ,
Park CM. 2009. The MYB96 Transcription factor mediates abscisic
acid signaling during drought stress response in Arabidopsis. Plant
Physiology, 151: 275–289.
Stracke R, Werber M, Weisshaar B. 2001. The R2R3-MYB gene family
in Arabidopsis thaliana. Current Opinion in Plant Biology 4: 447–456.
Takai R, Isogai A, Takayama S, Che FS. 2008. Analysis of flagellin percep-
tion mediated by flg22 receptor OsFLS2 in rice. Molecular Plant-
Microbe Interactions 21: 1635–1642.
Tang D, Simonich MT, Innes RW. 2007. Mutations in LACS2, a long-
chain acyl-coenzyme A synthetase, enhance susceptibility to avirulent
Pseudomonas syringae but confer resistance to Botrytis cinerea in Arabid-
opsis. Plant Physiology 144: 1093–1103.
Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G,
Katagiri F. 2003. Quantitative nature of Arabidopsis responses during
compatible and incompatible interactions with the bacterial pathogen
Pseudomonas syringae. Plant Cell 15: 317–330.
Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. 2005.
Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis
requires the MYB75 ⁄ PAP1 gene. Plant Physiology 139: 1840–1852.
de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield
JW, Rodriguez Egea P, Bo¨gre L, Grant M. 2007. Pseudomonas syringae
pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to
cause disease. EMBO Journal 26: 1434–1443.
Udvardi MK, Czechowski T, Scheible WR. 2008. Eleven golden rules of
quantitative RT-PCR. Plant Cell 20: 1736–1737.
482
Research
New
Phytologist
The Authors (2010)
Journal compilation New Phytologist Trust (2010)
New Phytologist (2010) 186: 471–483
www.newphytologist.com

Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan
N, Blundell TL, Esch JJ, Marks MD, Gray JC. 1999. The TRANSPAR-
ENT TESTA GLABRA1 locus, which regulates trichome differentiation
and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat
protein. Plant Cell 11: 1337–1350.
Wildermuth MC, Dewdney J, Wu G, Ausubel FM. 2001. Isochorismate
synthase is required to synthesize salicylic acid for plant defence. Nature
414: 562–565.
Winkel-Shirley B. 2001. Flavonoid biosynthesis. A colorful model for
genetics, biochemistry, cell biology, and biotechnology. Plant Physiology
126: 485–493.
Winkel-Shirley B. 2002. Biosynthesis of flavonoids and effects of stress.
Current Opinion in Plant Biology 5: 218–223.
Yasuda M, Ishikawa A, Jikumaru Y, Seki M, Umezawa T, Asami T, Mar-
uyama-Nakashita A, Kudo T, Shinozaki K, Yoshida S et al. 2008.
Antagonistic interaction between systemic acquired resistance and the
abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell
20: 1678–1692.
Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. 2003. A network of
redundant bHLH proteins functions in all TTG1-dependent pathways
of Arabidopsis. Development 130: 4859–4869.
Supporting Information
Additional supporting information may be found in the
online version of this article.
Fig. S1 Transcript abundances of the genes involved in
anthocyanin biosynthesis in the activation-tagging myb96-
1d line and myb96-1 mutant.
Fig. S2 Transcript abundances of the pathogenesis-related
genes in the activation-tagging myb96-1d line and the
myb96-1 mutant.
Fig. S3 Phenotypes of the myb96-1d X sid2 genetic cross.
Fig. S4 Transcript abundances of the RD22 gene in the
myb96-1d X Col-0 and myb96-1d X NahG genetic crosses.
Table S1 Primers used in this work
Please note: Wiley-Blackwell are not responsible for the
content or functionality of any supporting information sup-
plied by the authors. Any queries (other than missing mate-
rial) should be directed to the New Phytologist Central
Office.
New
Phytologist
Research
483
The Authors (2010)
Journal compilation New Phytologist Trust (2010)
New Phytologist (2010) 186: 471–483
www.newphytologist.com
Wyszukiwarka
Podobne podstrony:
REALIZM MORALNY, REALIZM MORALNY = tendencja dziecka do tego, by traktować obowiązki i związane z ni
Homoseksualiści są traktowani jako grupa pod specjalną opieką, Ruch Narodowy
Reformy szkolne są?finiowane jako istotny element infrastruktury społecznej
Mediator jako antropolog – podejĂcie antropologiczne w rozwiÈzywaniu konfliktów pracowniczych Leszek
Kwalifikacje zawodowe są rozumiane jako układ wiadomości
Mediator akademicki jako przeciwdzialanie mobbingowi w środowisku akademickim
Należności w sensie ekonomicznym rozumiane są jako zgodne z przepisami prawa
Reformy szkolne są definiowane jako istotny element infrastruktury społecznej docx
Epidemiologia jako nauka podstawowe założenia
4 socjalizacja jako podstawowy proces spoeczny
style poznawcze jako przykład preferencji poznawczych
radio jako medium audialne
na niebie są widoczne różne obiekty astronomiczne
POCH SA
socjologia jako nauka
Nakłucie prenatalne jako przyczyna krwotoku do jamy otrzewnej
Język jako narzędzie paradoksy
więcej podobnych podstron