
PCR i inne techniki
molekularne
Krzysztof Giannopoulos

Zastosowanie metod
wykrywania antygenu w
zależności od etapu
syntezy białka
•
DNA
•
RNA
•
Polipeptyd
•
Modyfikacja
wewnątrzkomórkow
a
•
Wydzielanie
•
Modyfikacja
zewnątrzkomórkowa
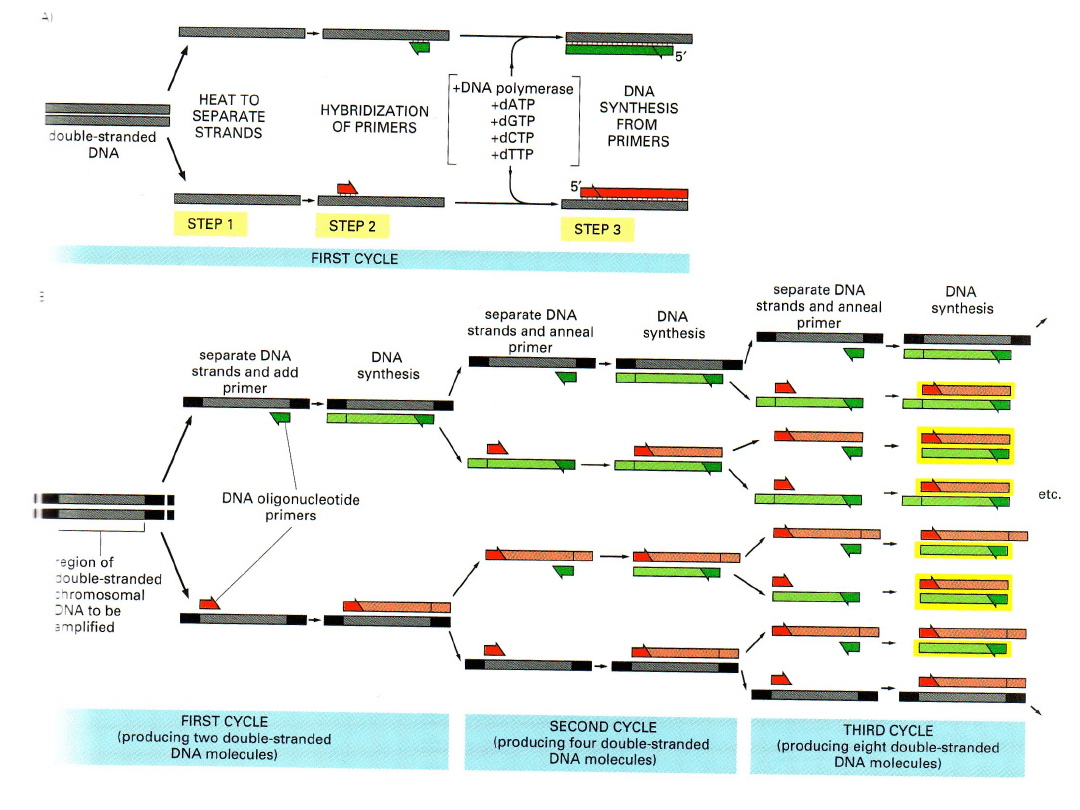
• PCR
• RT PCR
• Western blotting
• cytometria
wewnątrzkomórko
wa
• cytometria
przepływowa
• ELISA

Techniki analiz
genetycznych
• Southern blot (SB) analysis
• Polymerase chain reaction (PCR), and
real-time quantitative PCR (RQ-PCR)
• konwencjonalna cytogenetyka
• Fluorescence in situ hybridisation (FISH)
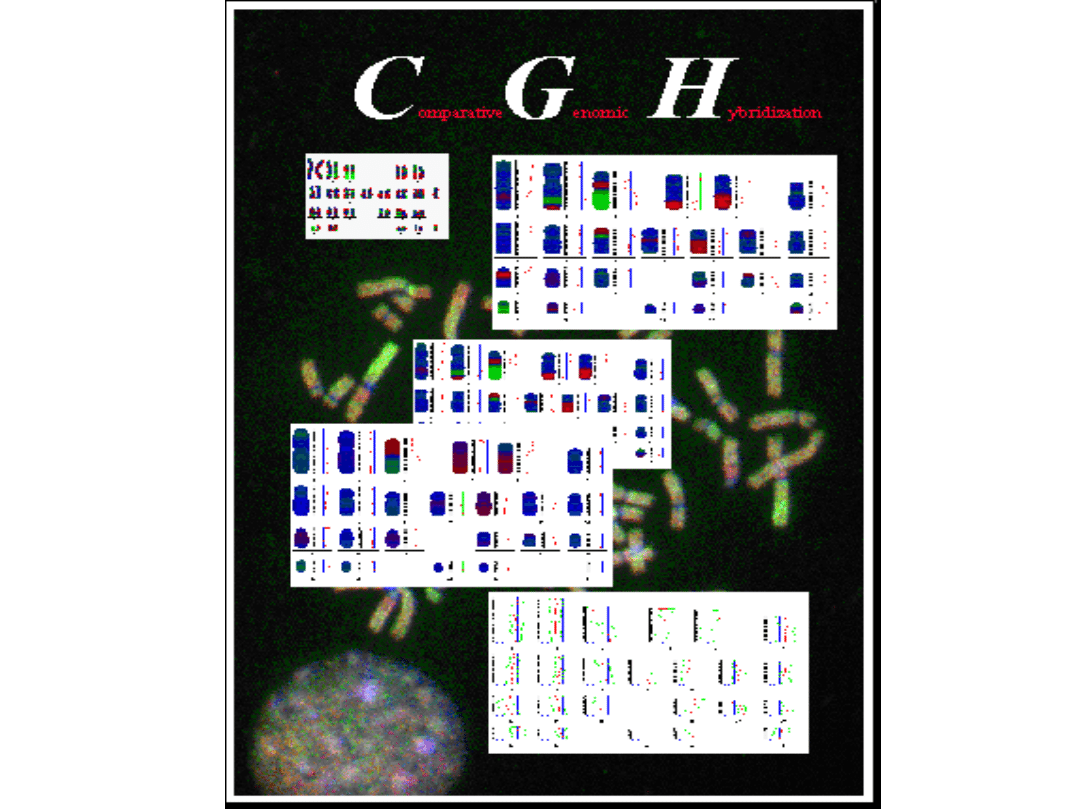
• comparative genomic hybridisation (CGH)
• gene expressions profiling (GEP)

TRI Reagent® is a single, homogenous solution for the isolation of total RNA. This
phenol-based reagent contains a unique combination of denaturants and RNase
inhibitors and is used in a convenient, single-step disruption/separation procedure.
The tissue or cell sample is homogenized or disrupted in the TRI Reagent, chloroform
is mixed with the lysate, and the mixture is separated into three phases by
centrifugation. The RNA is then precipitated from the aqueous phase with
isopropanol. The entire procedure can be completed in no more than one hour to
produce high yields of intact RNA for use in Northerns, nuclease protection assays, RT-
PCR and in vitro translation. TRI Reagent is especially effective for purifying RNA from
microorganisms.
Description: TRI Reagent is an improved version of the single-step total RNA isolation
reagent developed by Chomczynski (1). The RNA isolation method based on this
reagent is widely recognized and proven for RNA applications and is supported by a
substantial publication list (2,3,4,5). It is ideal for quick, economical, and efficient
isolation of total RNA or the simultaneous isolation of RNA, DNA and proteins from
samples of human, animal, plant, yeast, bacterial and viral origin.
References
1. Chomczynski, P. and Sacchi, N., Single-step method of RNA isolation by acid
guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156
(1987).
2. Chomczynski, P., A reagent for the single-step simultaneous isolation of RNA, DNA
and proteins from cell and tissue samples. BioTechniques 15: 532-537 (1993).
3. Mackey, K. and Chomczynski, P., Long-term stability of RNA isolation reagents. J. NIH
Res. 8: 72 (1996).
4. Wilfinger, W., et al., Effect of pH and ionic strength on the spectrophotometric
assessment of nucleic acid purity.BioTechniques 22: 474-481(1997).
5. Chomczynski, P. and Mackey, K., Modification of the Tri Reagent. procedure for
isolation of RNA from polysaccharide- and proteoglycan-rich sources.BioTechniques
19: 924-945(1995).
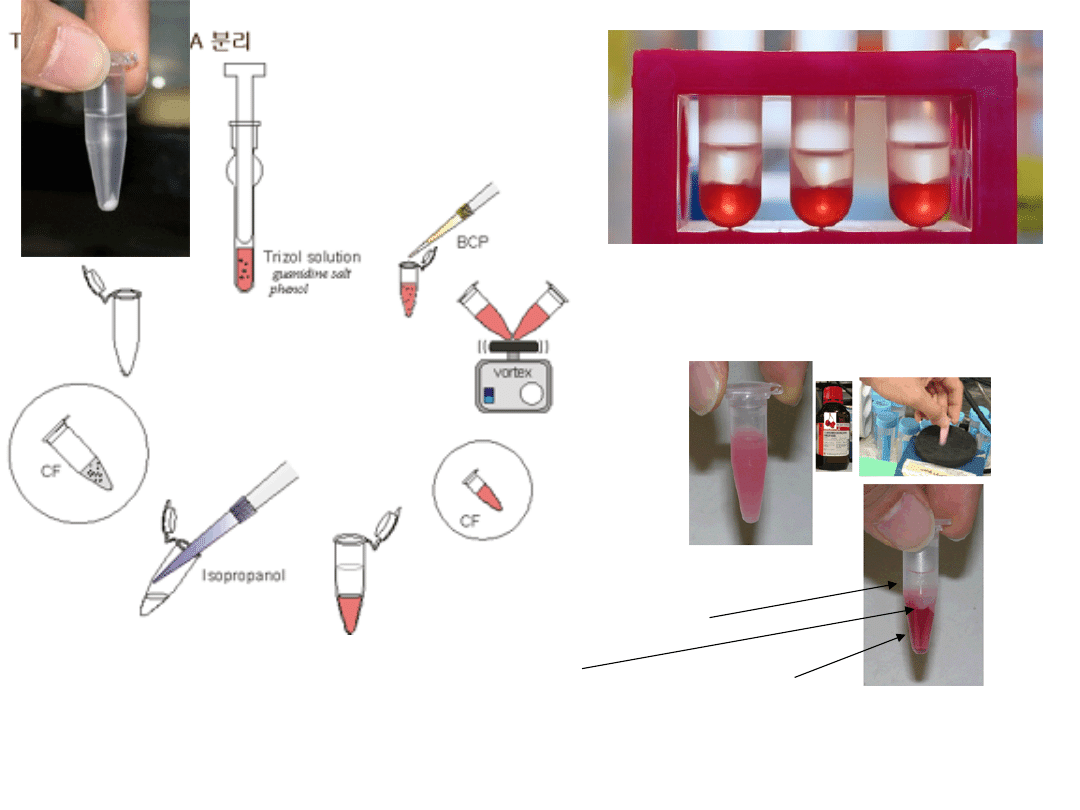
aqueous phase (RNA)
interphase
Phenol-chlorophorm (proteine + DNA)
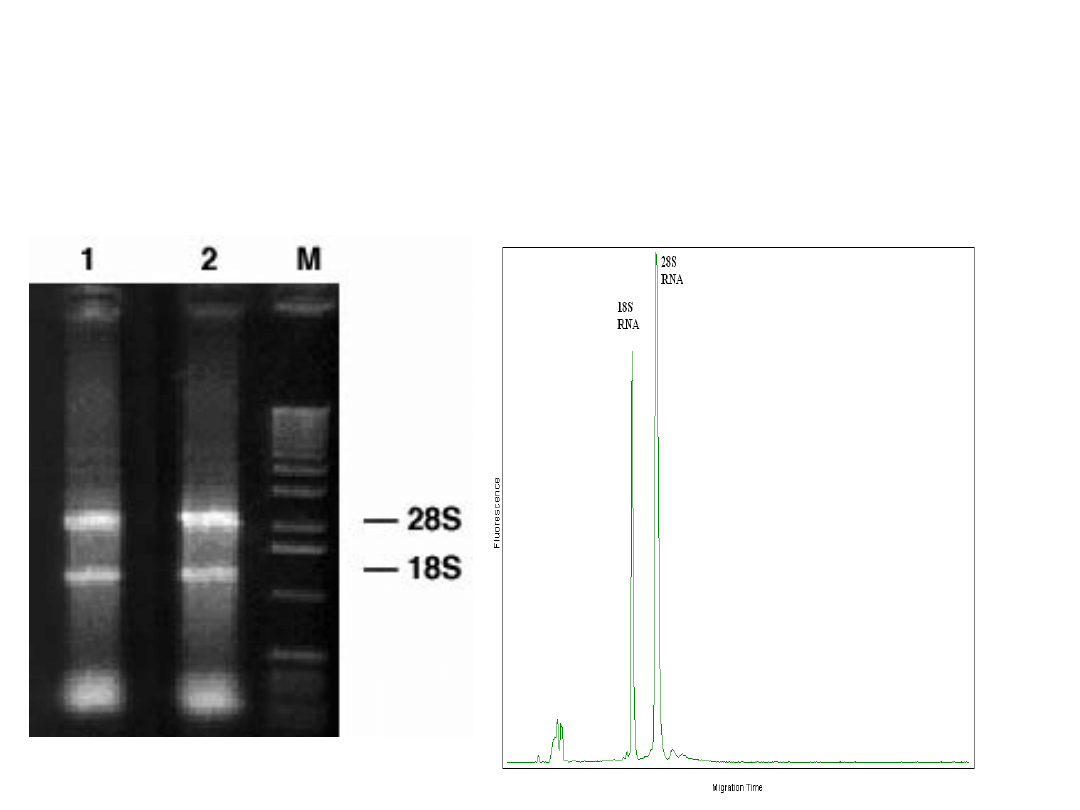
Jakość izolowanego RNA
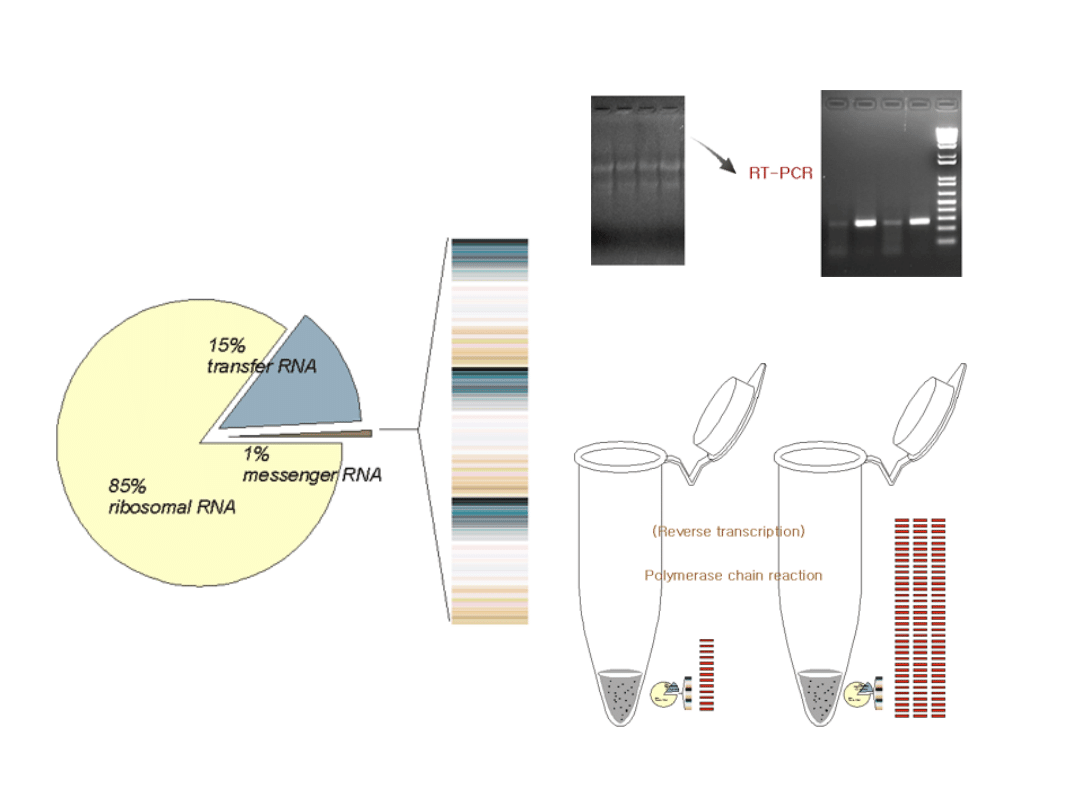
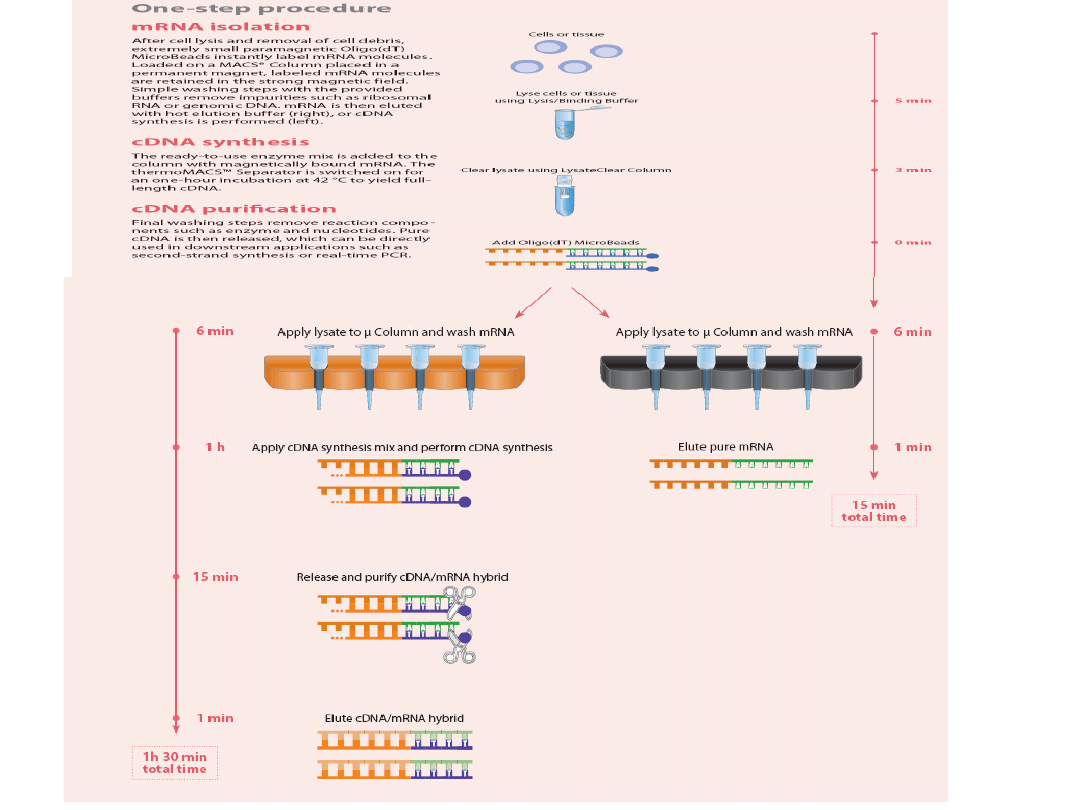
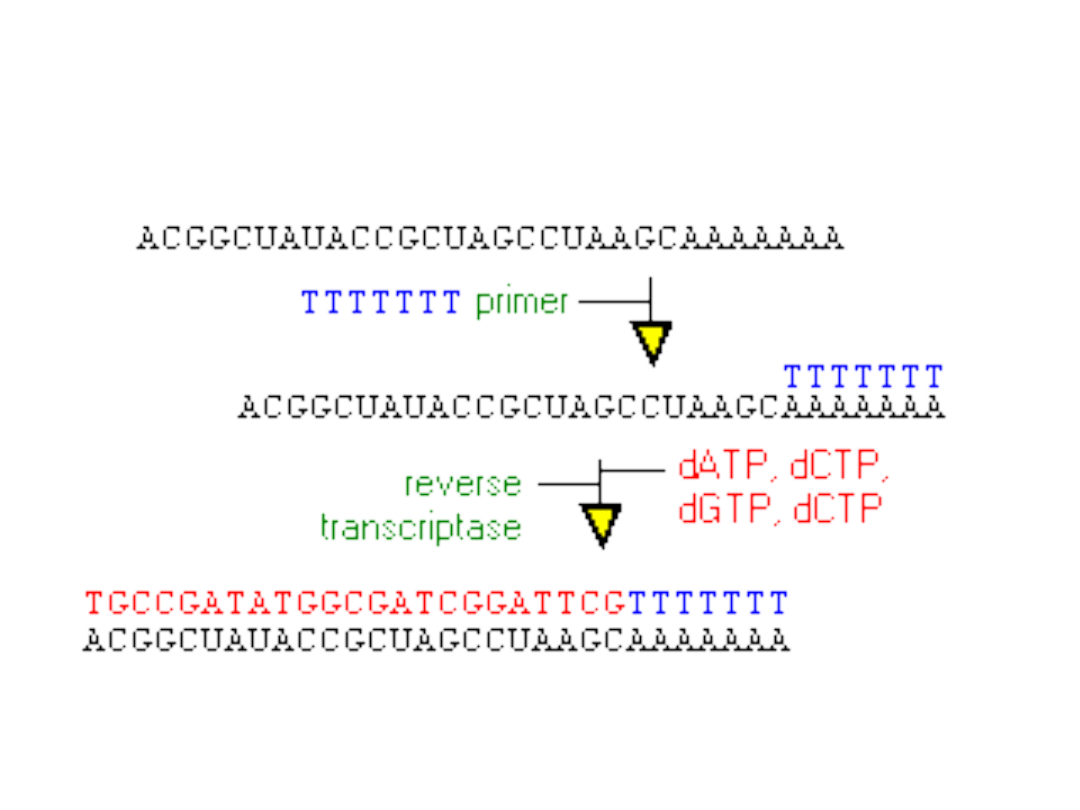
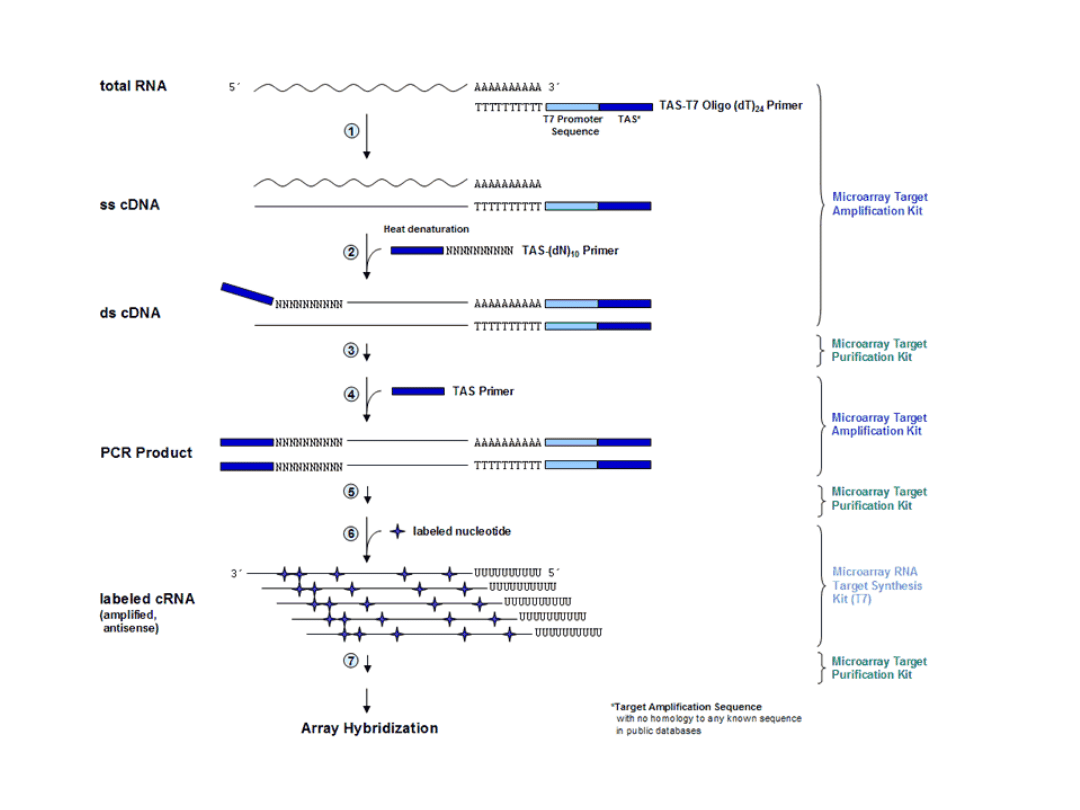
Reakcja odrwotnej transkrypcji
RT (reverse transcription)

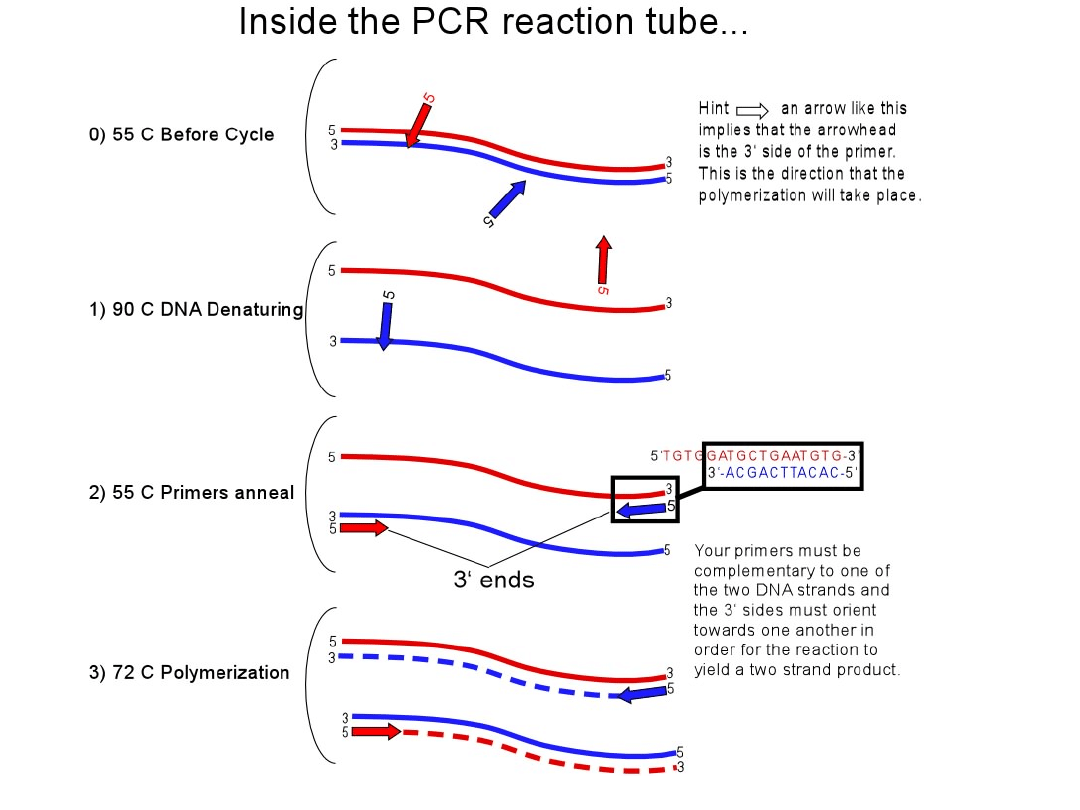
Rodzaje RT-PCR
• RT-PCR (reverse transcriptase polimeraze
chain reaction) reakcja lancuchowej
polimerazy z odwrotna transkrypcja
• qRT-PCR (quantitative RT-PCR) – ilosciowy
RT-PCR
• rtRT-PCR („real time“ RT-PCR) – ilosciowy
RT-PCR
• Competitive RT-PCR
• RT-PCR „in situ“
• RT-PCR muliplex

Quantify relative opsin gene
expression
• Make total RNA from retinas of single adult fish
–
Trizol
–
Quantify RNA by A260/A280
• Reverse transcribe 1 ug of total RNA using T
17
primer and
Superscript II
• Make master mix of PCR reagents for each fish
–
15 ul 2x Taqman ready made PCR mix
–
1 ul
RT reaction
–
5 ul water
– 21 ul
• Make primer / probe master mix for each gene of interest
–
3 ul forward primer (3 uM)
–
3 ul reverse primer (3 uM)
–
3 ul Taqman probe (2 uM)
–
30 ul total
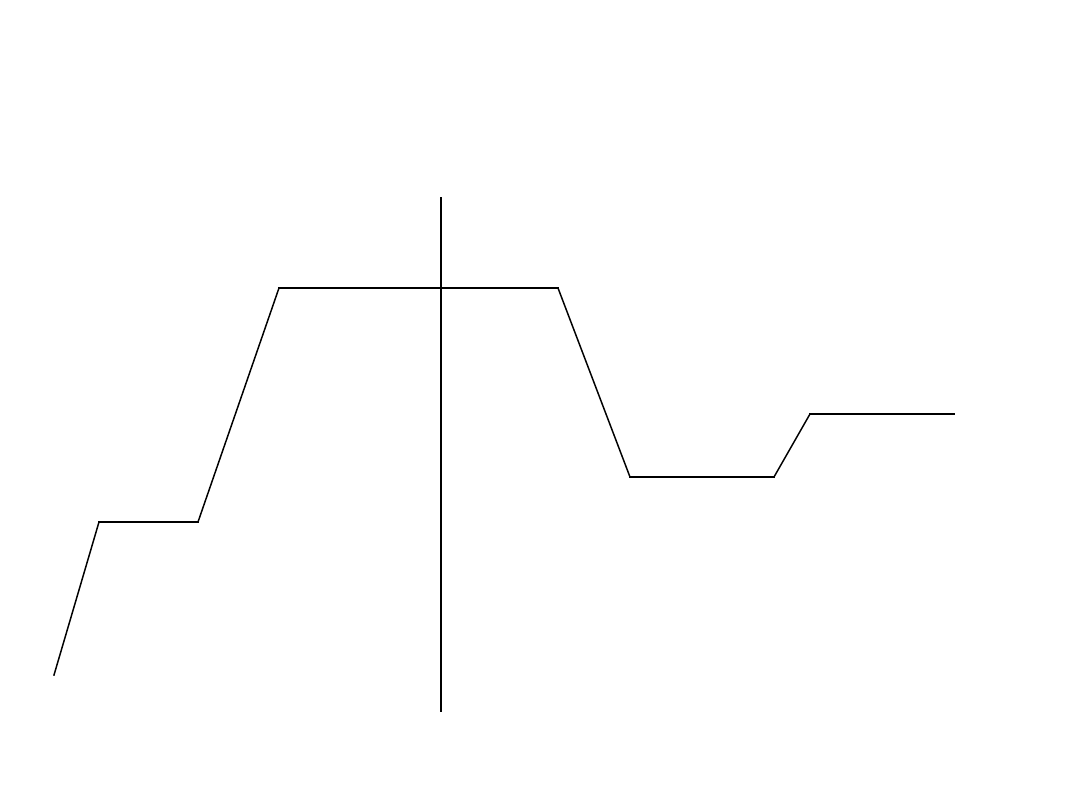
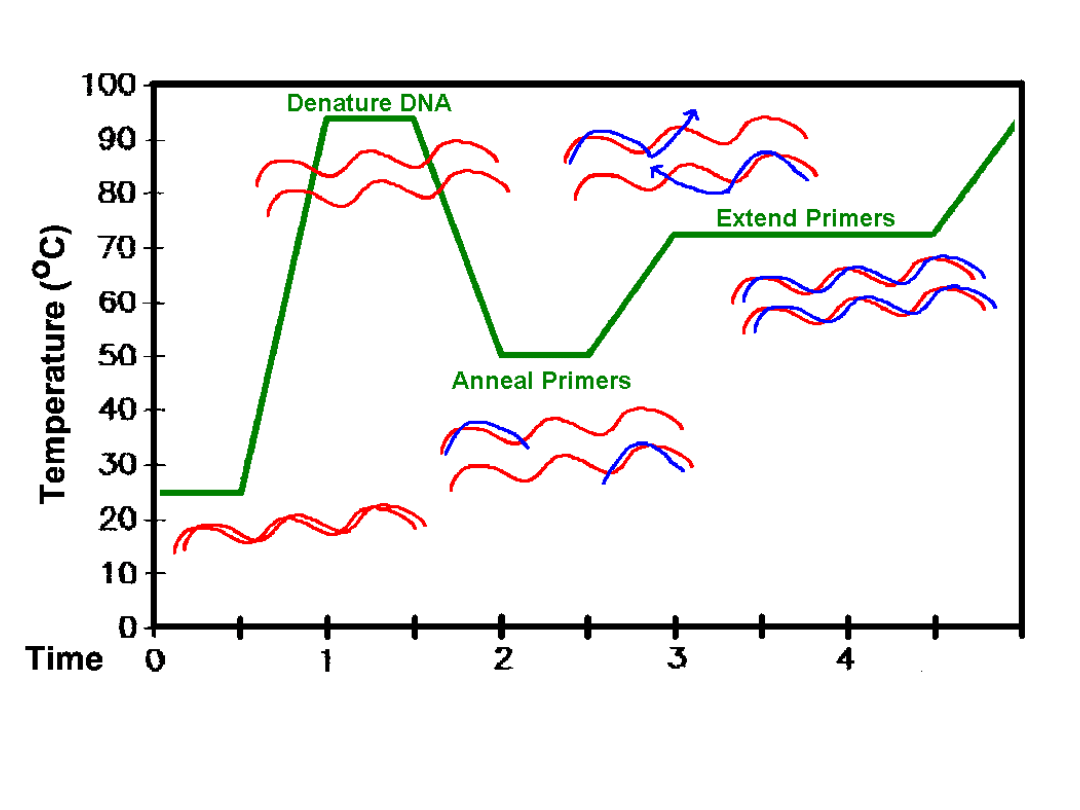
Run real time thermal cycle
50 C
5 min
95 C
10 min
20 s
55 C
30 s
65 C
1 min
40 cycles
Activates
UNG
enzyme
Activates
Amplitaq
Gold
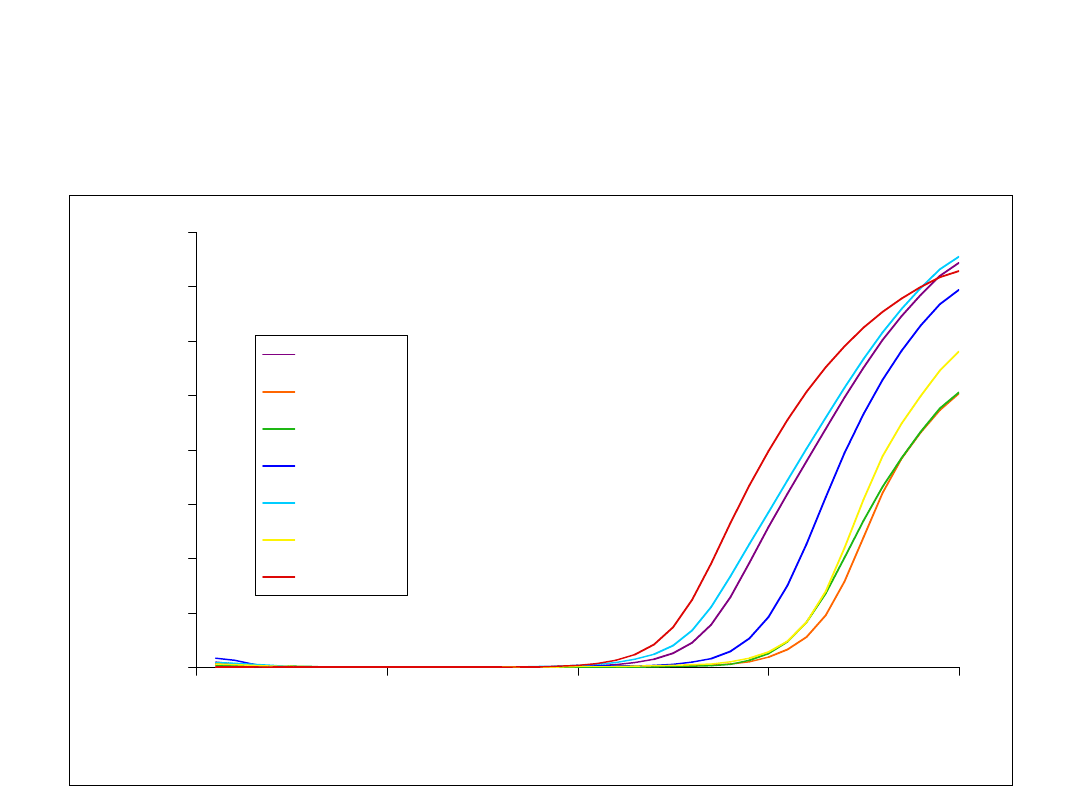
Screening BAC library for
red opsin gene
0
2
4
6
8
10
12
14
16
0
10
20
30
40
PCR cycle
R
n
SP5
SP6
SP7
SP8
SP9
SP10
Genomic
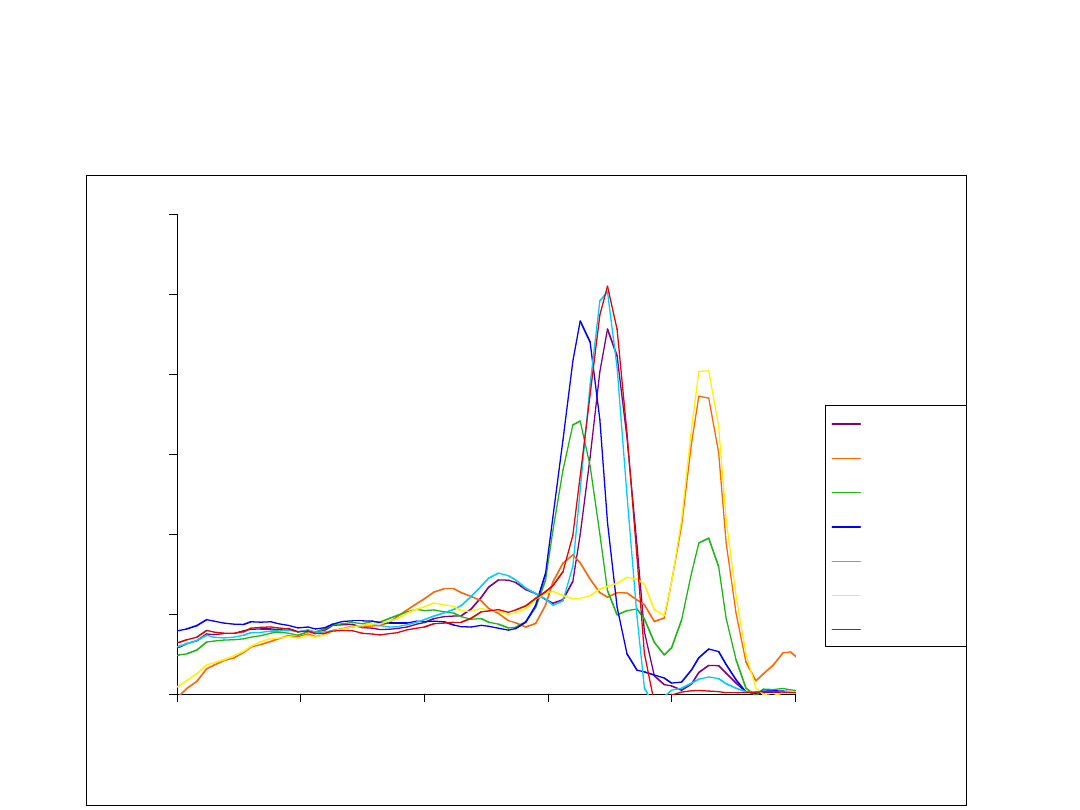
Dissociation curves: BAC
library screen for red opsin
0
1
2
3
4
5
6
65
70
75
80
85
90
Temperature
D
e
ri
v
a
ti
v
e
o
f
R
n
SP5
SP6
SP7
SP8
SP9
SP10
Genomic
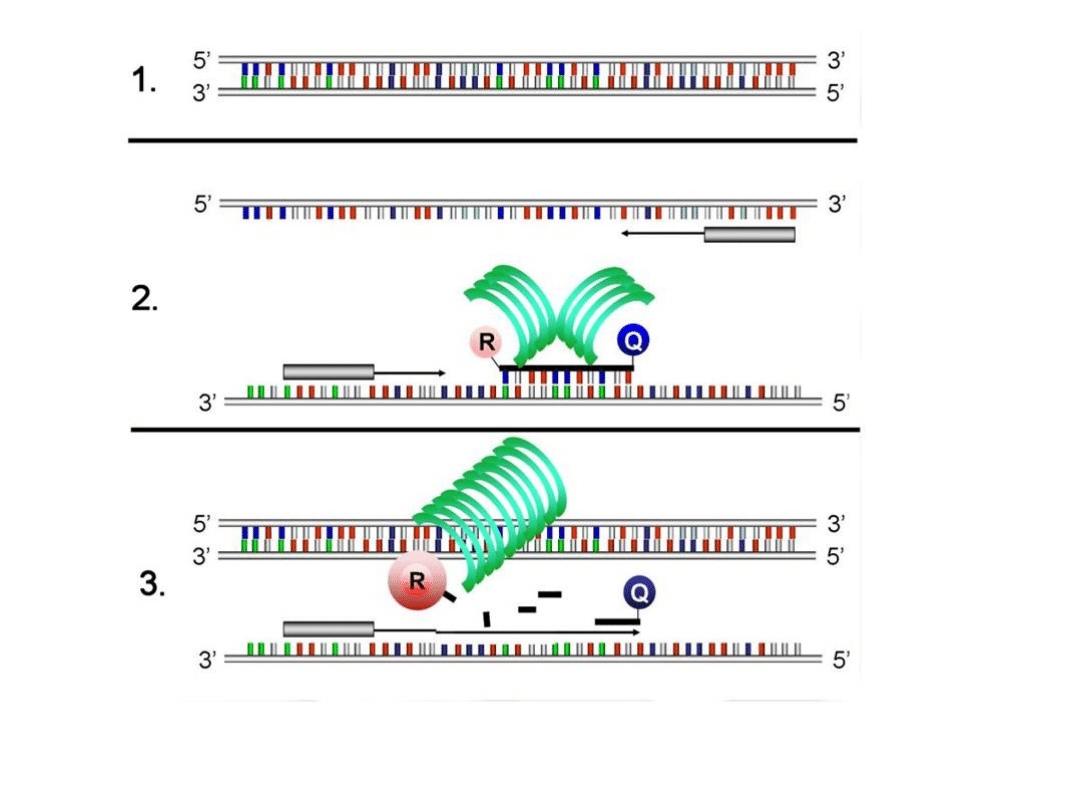
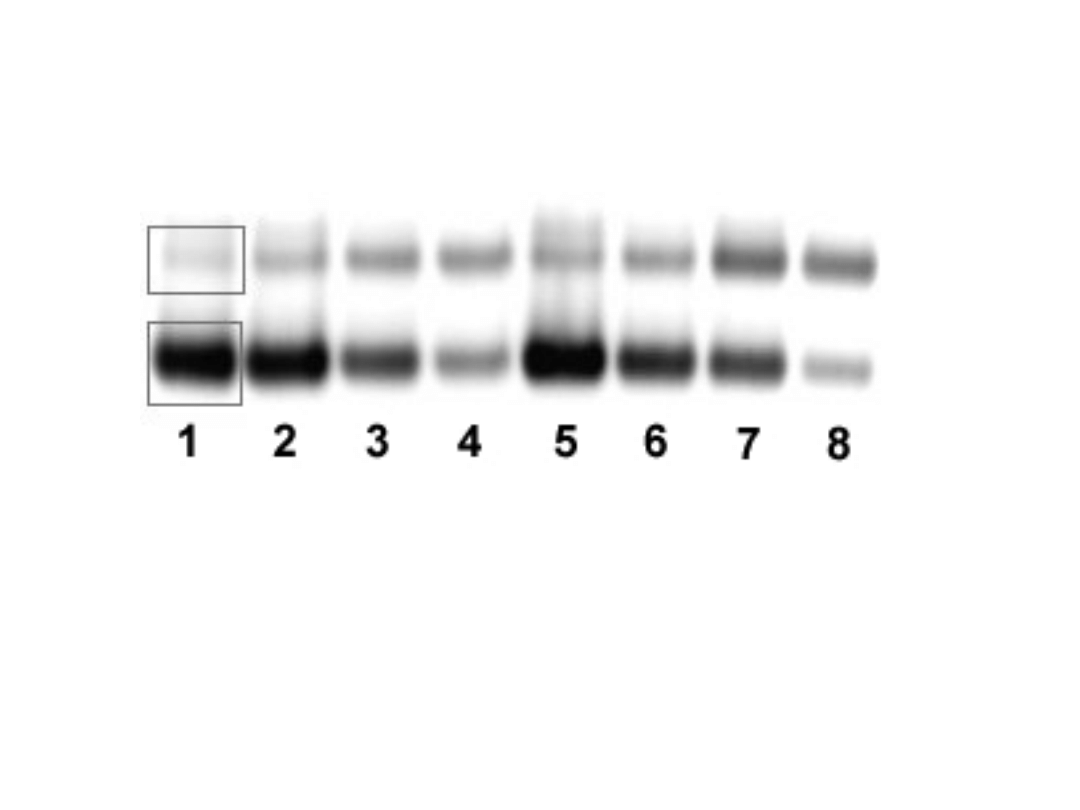
Competitive RT-PCR
Measurement of two RNA samples.
The lower bands are the standard bands, the upper bands are the
transcript bands. Lane 1-4: sample 3; lane 5-8: sample 4; lane 1+5: 250
pg standard; lane 2+6: 100 pg standard; lane 3+7: 50 pg standard; lane
4+8: 25 pg standard. The rectangles represent objects set with the
ImageQuant software.
Znacznik
wielkosci
Molecular
Weight Marker
VI

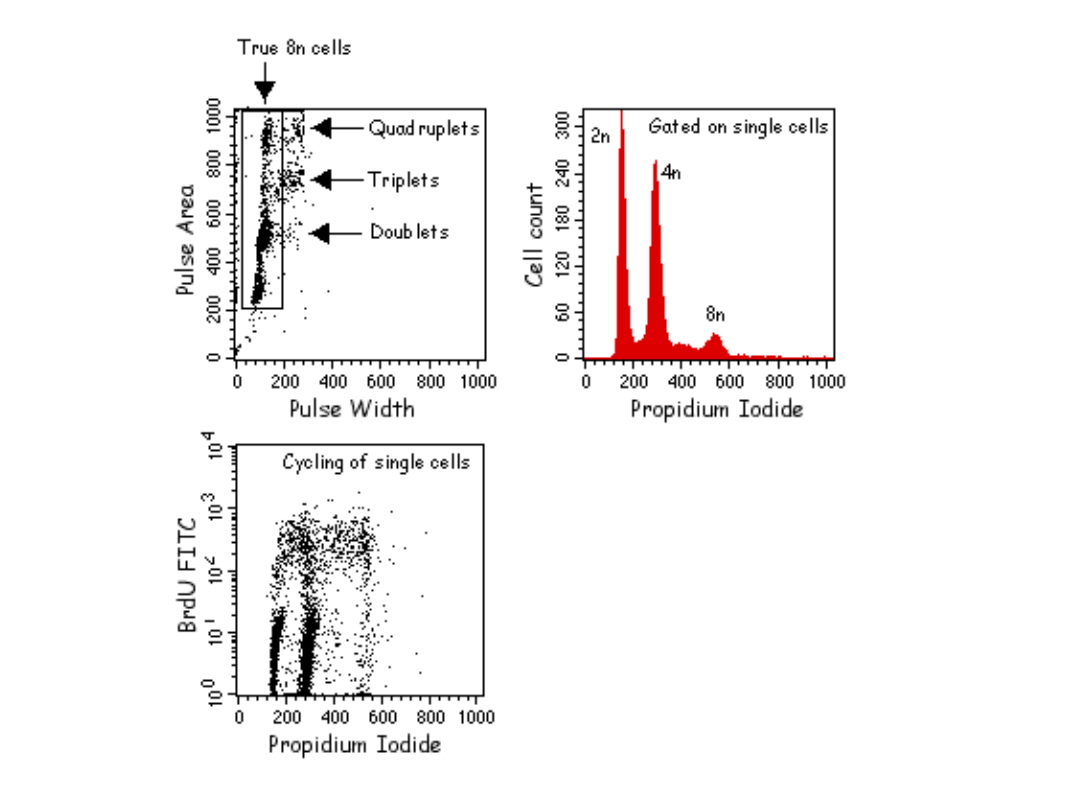
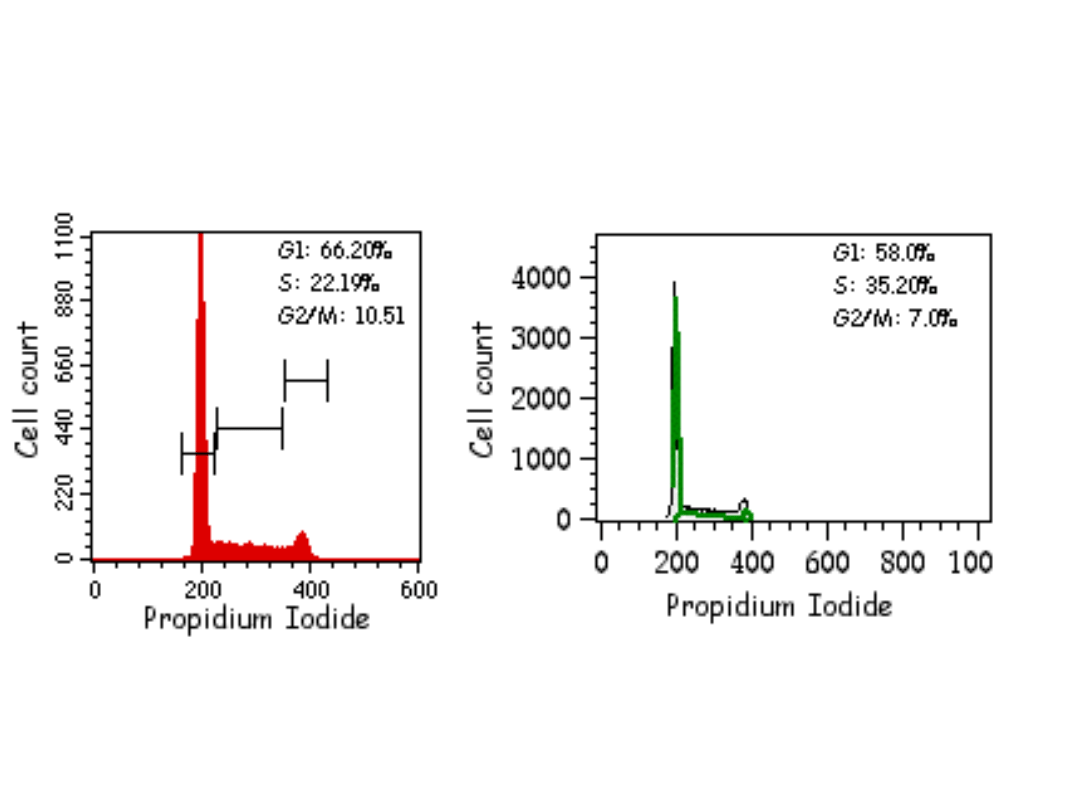
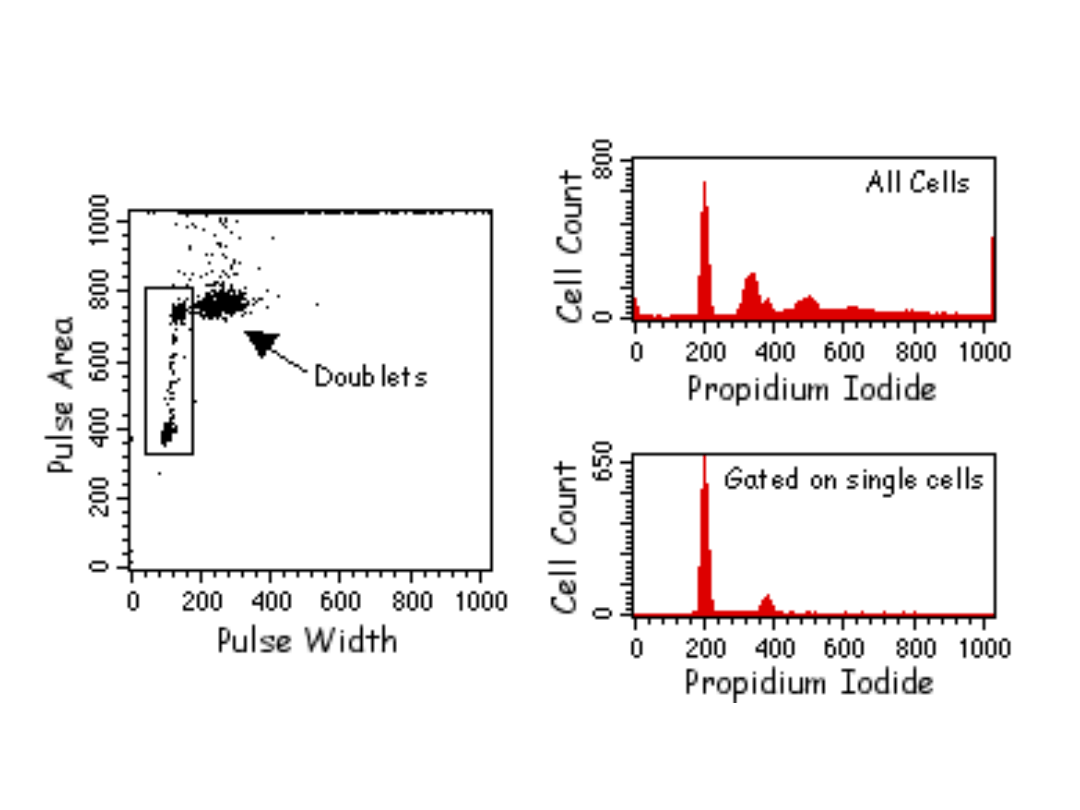
Znaczniki używane do analizy
cyklu komórkowego w
cytometrze przepływowym:
•UV excited: Hoechst 33342, Hoechst 33258, DAPI
•457nm excited: Mithramycin
•488nm excited: Propidium Iodide,
7Aminoactinomycin D, SYTOX Green, DRAQ5
•633nm excited: TO-PRO-3 Iodide
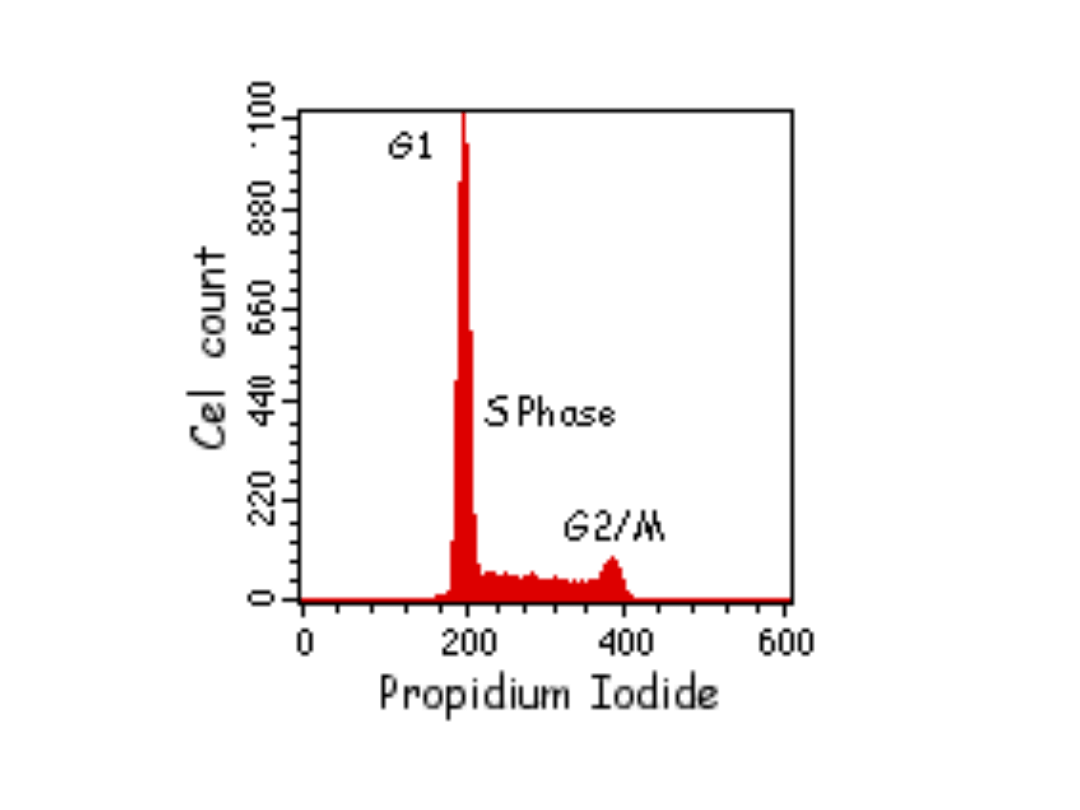

PROCEDURE:
•
Isolate cells:
•
1. For adherent cells, remove with PBS/EDTA and/or trypsin solution. For cells in
suspension, harvest by
•
centrifugation. Centrifuge cells at 1200 rpm at 4°C for 5 minutes, decant supernatant
and gently re-suspend
•
the cells in PBS.
•
2. Count the cells by hemocytometer.
•
3. Wash cells one time by putting 1X105 cells per tube, adding 1 ml of PBS and
centrifuging at 1200 rpm at
•
4°C. Re-suspend pelleted cells in 0.3 ml of PBS buffer.
•
Fixing the cells:
•
1. To fix the cells, gently add 0.7 ml cold ethanol (70%) dropwise to tube containing
0.3 ml of cell suspension in PBS while vortexing gently.
•
2. Leave on ice for 1 hour (or up to a few days at 4°C).
•
3. Centrifuge cells as above, wash 1 time with cold PBS and re-centrifuge.
•
4. Re-suspend cell pellet in 0.25 ml of PBS, add 5 µl of 10 mg/ml Rnase A (the final
concentration being
•
0.2-0.5 mg/ml).
•
5. Incubate at 37°C for 1 hour.
•
6. Add 10 µl of 1 mg/ml PI solution (the final concentration being 10µg/ml). Keep in
the dark and at 4°C until
•
analysis.
•
7. Analyze on FACS by reading on cytometer at 488 nm.
•
Propidium iodide stain (PI): Make a 1 mg/ml solution of PI in deionized
water.
Document Outline
- Slide 1
- Slide 2
- Slide 3
- Slide 4
- Slide 5
- Slide 6
- Slide 7
- Slide 8
- Slide 9
- Slide 10
- Slide 11
- Slide 12
- Slide 13
- Slide 14
- Slide 15
- Slide 16
- Slide 17
- Slide 18
- Slide 19
- Slide 20
- Slide 21
- Slide 22
- Slide 23
- Slide 24
- Slide 25
- Slide 26
- Slide 27
- Slide 28
- Slide 29
- Slide 30
- Slide 31
Wyszukiwarka
Podobne podstrony:
genetyka, ćw 6 geny, 6 Techniki oparte na PCR do diagnozowania chorów genetycznych i uchwycenia zmie
PCR RAPD Genetyka molekularna ćw koło 3
Ćw 4 Elektroforeza wyników PCR
Ćw 3 Identyfikacja płci u ptakow metoda PCR
Ćw 3 Identyfikacja płci u ptakow metoda PCR
Wyniki PCR ćw. gr. I 2012, Studia, I semestr III rok, Biologia molekularna
cw 12 pcr
cw RT PCR
ćw 4 Profil podłużny cieku
biofiza cw 31
Kinezyterapia ćw synergistyczne
Cw 1 ! komorki
W5 sII PCR i sekwencjonowanie cz 2
Pedagogika ćw Dydaktyka
Cw 3 patologie wybrane aspekty
Cw 7 IMMUNOLOGIA TRANSPLANTACYJNA
Cw Ancyl strong
więcej podobnych podstron