
Journal of Basic Microbiology 2011, 51, 357 – 363
357
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
Research Paper
Starvation survival of Candida albicans in various water
microcosms
Kamel Chaieb*
,
1
, Bochra Kouidhi*
, 1
, Tarek Zmantar
1
, Kacem Mahdouani
2
and Amina Bakhrouf
1
1
Laboratoire d’Analyses, Traitement et Valorisation des Polluants de l’Environnement et des Produits,
Faculté de Pharmacie, Université de Monastir, Tunisie
2
Laboratoire de Biologie Moléculaire, Hôpital Régionale de Kairouan, Tunisie
Candida is a major Human pathogen causing a variety of infections and can survive for ex-
tended period of time in aquatic environment including marine and fresh water. In this study
we compared a colorimetric XTT assay to colony forming units (CFU) count to evaluate the
survival potential of Candida albicans incubated in water microcosms. Our results showed that
cells maintain cultivability within a long period followed by a decline in cultivability and a
drop of plate counts to less than 20 cell ml
–1
after 150 days in tap water, 190 days in rain water
and 200 days in seawater. In addition we noted that 10% of cells viability was reached after
150 days in seawater, 180 days in rain water and 210 days in tap water. Molecular method
confirms the persistence of C. albicans cells in water during long time starvation period.
Keywords: Candida albicans / Water / CFU / XTT / Its
Received: August 02, 2010; accepted: December 13, 2010
DOI 10.1002/jobm.201000298
Introduction
*
Candida is a major Human fungal pathogen causing a
variety of infections ranging from superficial mucosal
diseases to deep seated mycoses [1]. It can cause super-
ficial skin, vaginitis, and nosocomial infections in com-
promised hosts [2]. Enumeration and characterization
of yeasts have been reported for marine [3, 4], estuaries
[5] and fresh waters [6]. A high density of C. albicans has
been found to be associated with recent human fecal
contamination in temperate fresh waters [7]. A wide
occurrence of fungi in Norwegian drinking water was
also reported [8]. Pereira et al., [9] found that Candida,
Cryptococcus and Kloeckera genus were the most fre-
quently detected yeasts in spring water, surface water,
and ground water, respectively. Stress responses in
fungi and the relationships between stress and fungal
virulence has been studied elsewhere [10]. Microorgan-
isms are often exposed to rapid variations in the quality
* Kamel CHAIEB
and Bochra KOUIDHI
were contributed equally in this
manuscript.
Correspondence: Kamel Chaieb, Laboratoire d’Analyses, Traitement et
Valorisation des Polluants de l’Environnement et des Produits, Faculté
de Pharmacie, Rue Avicenne 5000, Université de Monastir, Monastir,
Tunisie
E-mail: chaieb_mo@yahoo.fr
Phone: +21673461000
Fax: +21673461830
and availability of nutrients. For instance, the ability to
maintain a high catabolic machinery during starvation
has been studied in S. cerevisiae [11]. Further study
showed a long-term survival of C. albicans in distilled
water for 3–10 years [12–14]. Viability and morpho-
logical stability of a large number of yeast and fungi in
water has been reported for a period between 1 to
20 years [15].
Metabolic assays that rely on reduction of a redox
reactive dye (XTT) have been widely accepted for anti-
fungal susceptibility testing [16] and biofilm formation
[17]. XTT was reduced by the dehydrogenase enzymes
present in the electron transport system (ETS) to a wa-
ter soluble formazan dye and the absorbance can be
measured by spectrophotometry [18]. The intracellular
reduction of XTT releases a formazan compound easily
quantified by colorimetric estimation [19] and can be
adapted for in situ testing [20, 21].
The standard method for quantification of viable
C. albicans is the enumeration of colony forming unit
(CFU). Few studies have compared metabolic assay with
direct CFU counts for enumeration of viable C. albicans
[16, 17].
In the present study we examined the survival of C.
albicans in three water microcosms (sea water, tap water
and rain water) using CFU counting and XTT colorimet-

358 Kamel
Chaieb
et al.
Journal of Basic Microbiology 2011, 51, 357 – 363
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
ric reduction assay. In addition molecular detection by
PCR was assessed to confirm the persistence of C. albi-
cans cells during starvation period.
Materials and methods
Water quality
Three water microcosms were used in this study: sea-
water, tap water and rain water which were sterilized
through a 0.22 μm pore size filter.
The pH was measured with a digital pH meter, Inolab
(WTW, Germany). Dissolved oxygen was measured with
oxy-meter (oxi cal-ST, WTW, Germany). A model HACH
company (Loveland, CO, United States) was used to
measure conductivity and salinity. Turbidity and hard-
ness measurements were done in the field, using a
Laboratory Turbidimeter (Model 2100AN, HACH, CO,
United States). Calcium, iron, magnesium, chloride
(Cl
−
), sodium (Na
+
) and potassium (K
+
) were determined
using a Cobas Integra 400 plus analyzer (Roche Diag-
nostics, Belgium).
Candida albicans strain and growth conditions
Oral isolate of C. albicans were used in this study. The
identification was assessed using the Api ID32C system
(bioMérieux, Marcy l’Étoile, France) according to the
manufacturer′s recommendations and the results were
read using an automated microbiological mini-Api
(bioMérieux, Marcy l’Étoile, France).
Survival assay
The tested strain was grown in 100 ml Sabouraud broth
during 24 h at 30 °C. Cells were collected by centrifuga-
tion at 12,000 g for 15 min at 4 °C. The pellet was
washed three times with phosphate-buffered saline
(7 mM Na
2
HPO
4
, 3 mM NaH
2
PO
4
and 130 mM NaCl at
pH 7.4) and inoculated (2 ml) to 250 ml glass bottle con-
taining respectively 200 ml of sterile seawater, tap water
and rain water until a final concentration 10
6
colony
forming units (CFU) per ml was reached. Three flasks
containing 200 ml of sterile seawater, tap water and
rain water respectively were used as a negative control.
Three replicate flasks were used for each treatment.
All microcosms were incubated in a static state at am-
bient temperature (25 °C).
Enumeration techniques
Microcosms were sampled daily during the first week,
weekly during the first month, and then once a month
during eight months. Cultivable cells were determined
by the drop plate method [22, 23] using Sabouraud agar
plates. Time zero (inoculation time) and subsequent
samples were taken for plate counts. The plates were
incubated at 30 °C, and the number of CFU was deter-
mined after 24 h at 30 °C.
XTT colorimetric assay
XTT (Sigma-Aldrich Corp) was dissolved in PBS at a final
concentration of 1 mg/ml and sterilized through a
0.22 μm pore size filter. Menadione solution (Sigma-
Aldrich Corp) at 1 μM was used as an electron coupling
agent and prepared in acetone immediately before use.
Kinetic data were quantified biochemically using XTT
assay. For each assay a 100 μl aliquot of the XTT me-
nadione solution mixed at a volume ratio of 12.5:1 (v/v)
were added to 900 μl of cell suspensions from each
sample in Eppendorf tube. Then the tubes were incu-
bated in the dark at 30 °C for 3 h. The colorimetric
changes were measured at 492 nm using a spectropho-
temeter Spectro UV-Vis (Model UVD-2960, Labomed,
inc, California). All results were presented as mean
oxidative activity from three independent experiments
± standard deviation.
Percent reduction in formazan produced was calculat-
ed using the following formula: 100% – (Experimental
well absorbance at 492 nm – Blank absorbance at 492 nm)
× 100/Negative control absorbance at 492 nm [14].
Detection by PCR of Candida albicans
The presence of Candida albicans cells in various micro-
cosms was confirmed by PCR using Its86 5′-GTGAAT
CATCGAATCTTTGAAC-3′ and Its4 5′-TCCTCCGCTTATT
GATATGC-3′ primers [20]. Chromosomal DNA was ex-
tracted using a Wizard Genomic purification Kit
(Promega, USA) according to the Manufacturer's rec-
ommendation. PCR was performed
in a 25 μl reaction
volume containing: 20 ng of extracted DNA, 5 μl green
Go Taq buffer (5×), 200 μM of
each deoxynucleoside
triphosphates (dNTP), 0.5 μM
of each forward and re-
verse primer, 1 U of GO Taq DNA polymerase (Promega,
USA). Each PCR was performed twice for
confirmation
of the results. DNA extraction from the non inoculated
bottles was served as negative control.
PCR products (5 μl) were analyzed on 1% agarose gel
in 1X Tris-borate-EDTA buffer (TBE) pH 8.3, and visual-
ized under ultraviolet transillumination, photographed
using Gel Doc XR apparatus (Biorad, USA) and their
sizes were determined with 100 bp molecular size
marker.
Statistical analysis
Each analysis was performed using the SPSS 17.0 statis-
tics package for Windows. The differences in percent
Journal of Basic Microbiology 2011, 51, 357 – 363
Starvation survival of C. Albicans in water microcosms
359
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
Table 1. Water quality of the three studied microcosms.
Parameters
Unit
Sea water
Tap water
Rain water
Temperature
°C 28±3
28±3
28±3
pH
8.26
7.88
7.43
Dissolved oxygen (DO)
mg l
–1
6.47 6.6
6.10
Chloride (Cl
−
) g
l
–1
26.62
1.24
0.35
Salinity g
l
–1
34.4
1,006
0.114
Conductivity micosiemens/cm
56,000
2,000
240
Turbidity
NTU
1.05
1.16
1.32
Na+
mmol l
–1
524.3 9 2.8
K+
mmol l
–1
11.1 0.09
0.05
Calcium (Ca
2+
) mmol
l
–1
14.36 2.76
0.5
Iron
µ
mol l
–1
1.2 0.7 0.4
Magnesium (Mg
2+
) mmol
l
–1
12.94 1.6 0.03
Density 1.025
1.005
1.005
viability and the degree of biofilm formation as a func-
tion of starvation period were examined by the Fried-
man test, followed by the Wilcoxon signed ranks test.
P-values <0.05 were considered significant.
Results
Water quality
As presented in Table 1, water quality was quite vari-
able. Available nutrient was higher in seawater than
tap and rain water. Dissolved oxygen concentration
ranged from 6.47 mg l
–1
(sea water) to 6.10 mg l
–1
(rain
water). The pH values ranged from 8.26 to 7.43 in the
three studied microcosms (Table 1).
Our data revealed also that the concentration of so-
dium, chloride, calcium and magnesium was higher in
sea water in comparison to tap water and rain water.
In addition salinity in sea water had typical values
(26.62 g l
–1
). Moreover, conductivity values were higher
in sea water in comparison with the two remaining
microcosms.
We noted also that some ionic species such as Cl
–
and
Mg
2+
had a high level in seawater. However Ca
2+
and
Mg
2+
were quite low in tap and rain water.
Survival curves during starvation
In this study, the survival of C. albicans in various mi-
crocosms at ambient temperature under nutrient
starvation was investigated using CFU and XTT assay.
C. albicans incubated in various microcosms at room
temperature (mean 26 °C ± 3), remained cultivable for
prolonged time periods in all microcosms (Figs. 1, 2
and 3).
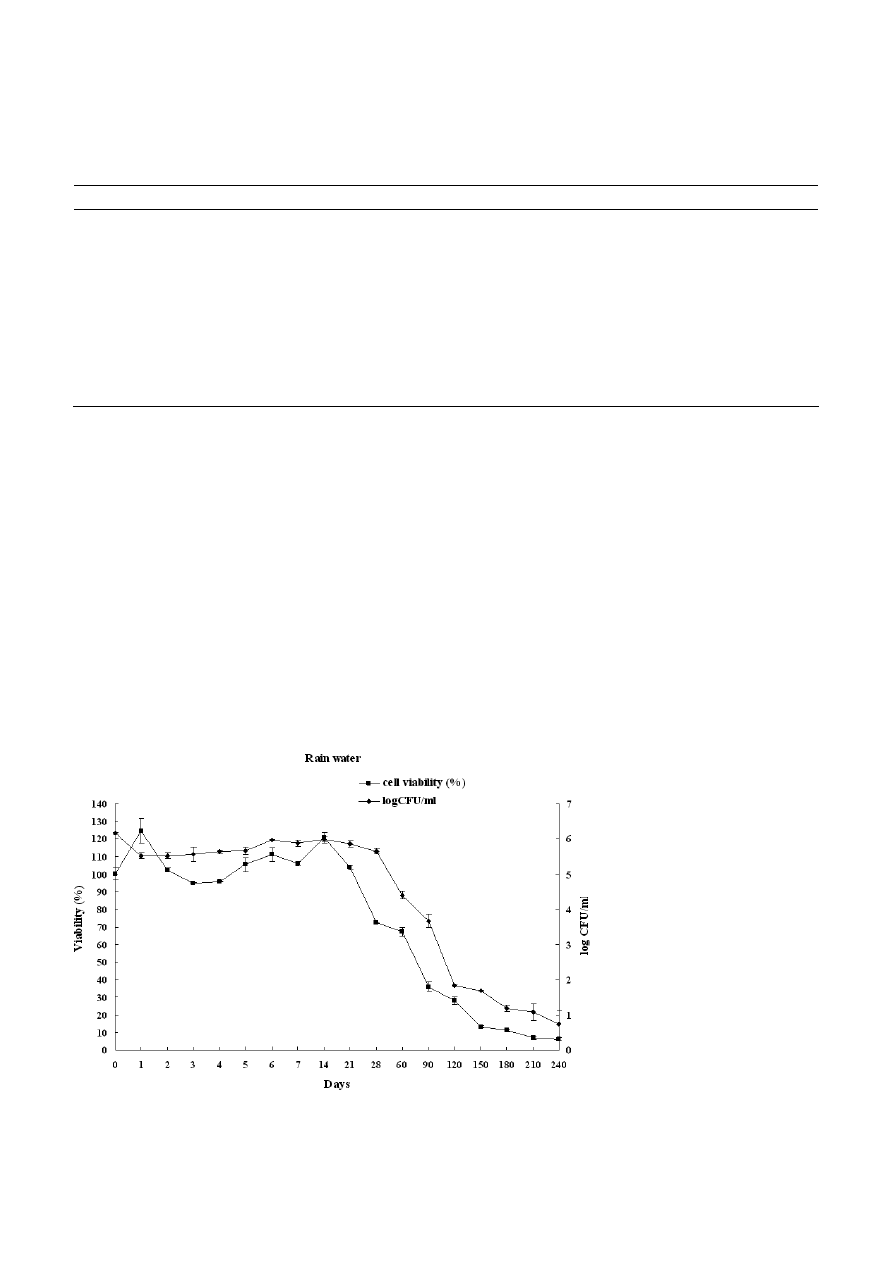
Figure 1. Survival curves of Candida albicans incubated in rain water determined by XTT and CFU assay after starvation period.
360 Kamel
Chaieb
et al.
Journal of Basic Microbiology 2011, 51, 357 – 363
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
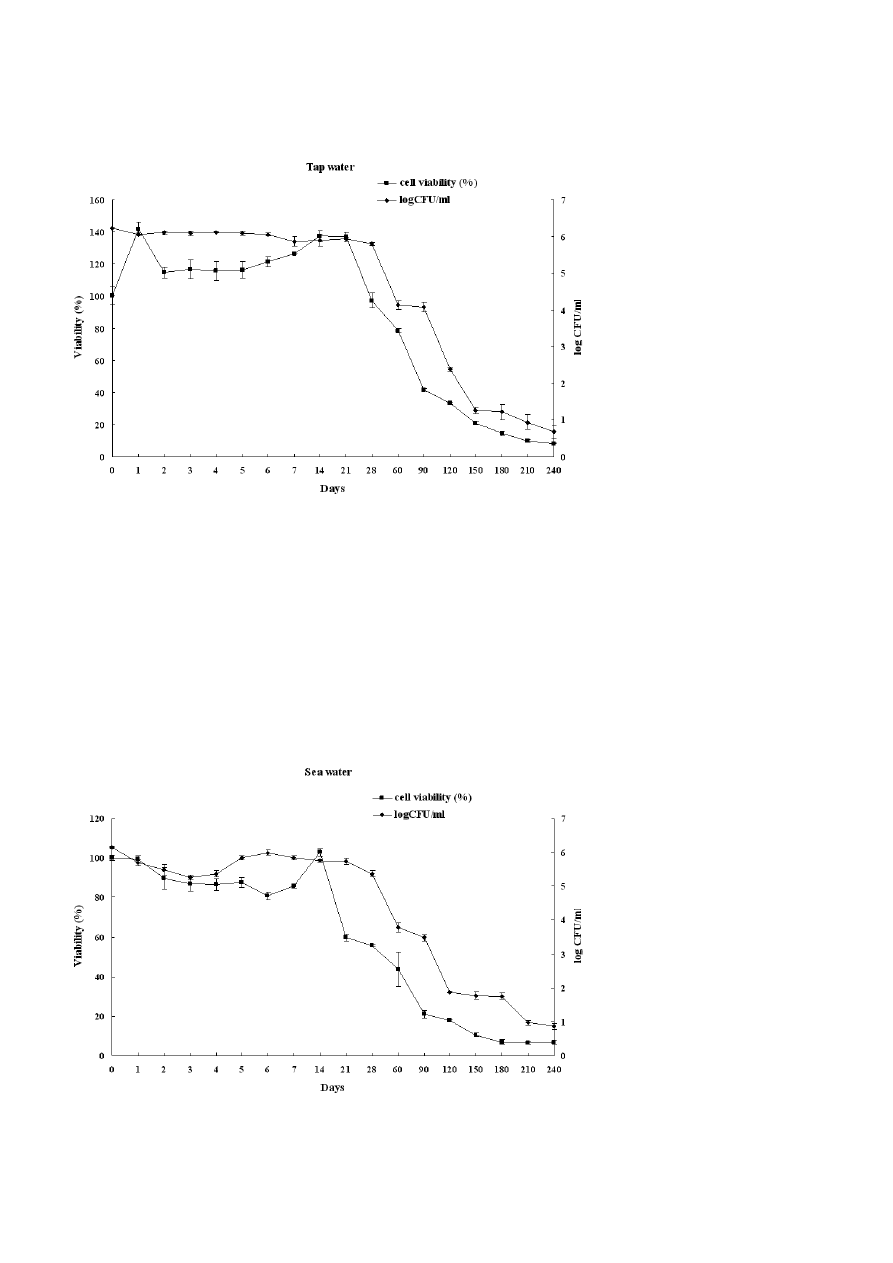
Figure 2. Survival curves of Candida albicans incubated in Tap water determined by XTT and CFU assay after starvation period.
During starvation period, the survival curves fell into
three stages: at the beginning of starvation, cells main-
tain cultivability within 14 days for rain water, 21 days
for sea water and 28 days for tap water. This was fol-
lowed by a decline in cultivability and then a drop of
plate counts to less than 20 cell ml
–1
after 150 days in
tap water, 190 days in rain water and 200 days in sea
water. We noted also a decrease of 4 log in CFU count
after 120 days of starvation in rain water and sea water
in comparison to 130 days in tap water.
Within 3 months, post-inoculation in sea water mi-
crocosms, total C. albicans populations remained at ap-
proximately 56 cells ml
–1
. The Wilcoxon signed ranks
test revealed a statistical significant difference between
the log CFU obtained after starvation period and con-
trol (P = 0.008).
In another way, the XTT assay was performed to
evaluate the oxidative activity of C. albicans cells under
nutrient starvation. Our result demonstrated that vi-
ability (%) increased slightly at the beginning of the
experiments (first days) and decreased after four days
in tap water and rain water and six days in sea water
than the percent viability of cells decreased with star-
vation time in all microcosms.
Figure 3. Survival curves of Candida albicans incubated in Sea water determined by XTT and CFU assay after starvation period.
Journal of Basic Microbiology 2011, 51, 357 – 363
Starvation survival of C. Albicans in water microcosms
361
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
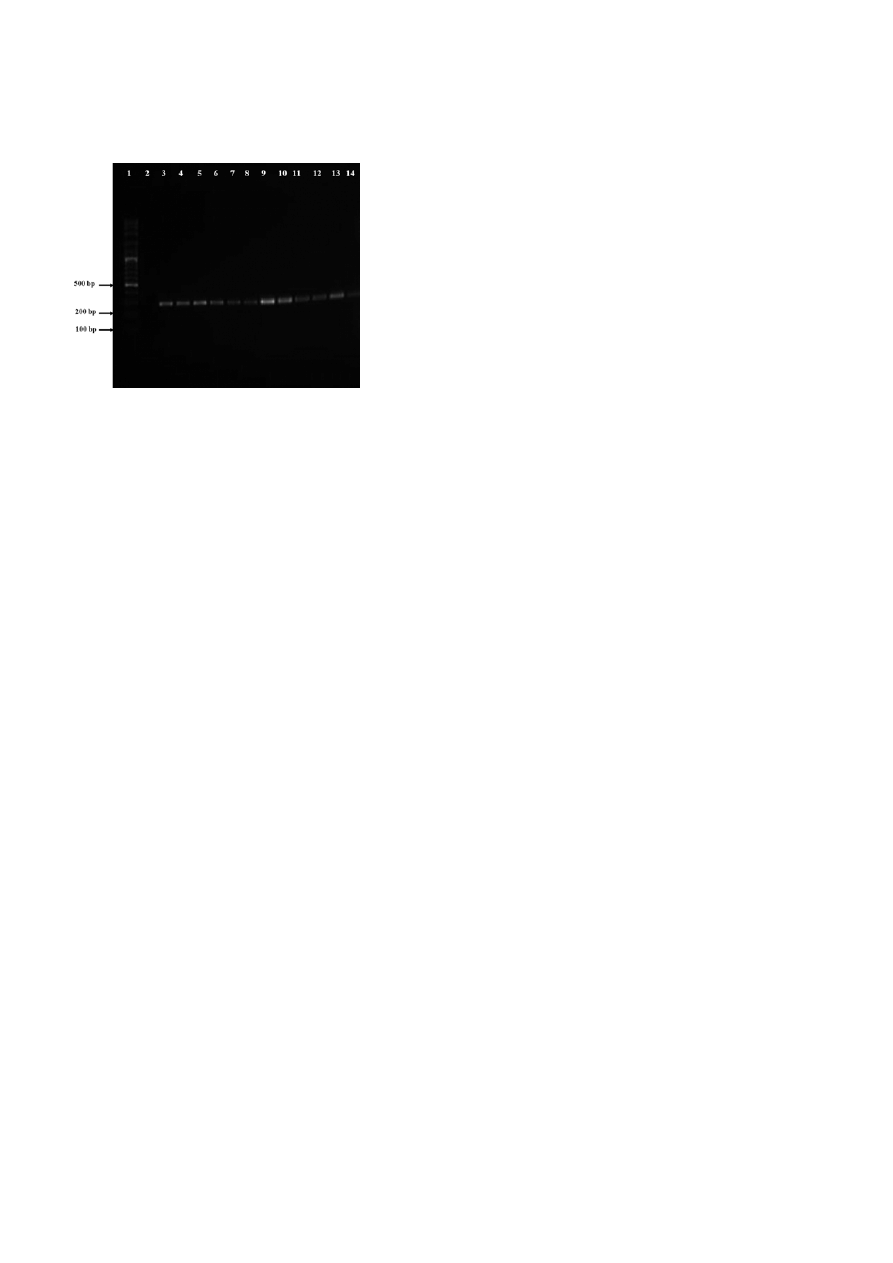
Figure 4. Agarose gel electrophoresis (1% agarose) of the ampli-
fication products obtained for starved Candida albicans strains.
Lanes was as follow: 1, molecular weight marker (100-pb DNA
ladder); 2, Negative control; 3 – 5 Candida albicans in microcosms
(Sea water, rain water and tap water respectively) after 1 days of
incubation period; 6 – 8 Candida albicans in water microcosms (Sea
water, rain water and tap water respectively) after 60 d of incubation
period; 9 – 11 Candida albicans in water microcosms (sea water,
rain water and tap water, respectively) after 180 d of incubation
period; 12 – 14 Candida albicans in water microcosms (sea water,
rain water and tap water respectively) after 240 d of incubation
period.
We noted also that 50% of cells viability was reached
after 45 days in sea water, 75 days in rain water and
80 days in tap water. In addition, metabolically active
cells measured by the XTT assay remained at around
10% after 150 days in sea water, 180 days in rain water
and 210 days in tap water (Figs. 1, 2 and 3).
Molecular detection of starved Candida albicans
As presented in Fig. 4, after starved period, all C. albi-
cans strains were successfully identified with poly-
merase chain reaction assay giving a 279 pb product
size [24].
Discussion
Several human pathogenic yeasts may persist as viable
organisms in natural environment and enter into the
so-called viable but non-culturable (VBNC) state. Lacks
of nutrients in natural ecosystem are often limiting
factor for microbial population. Viability of starved
cells has been largely studied in our laboratory for Sal-
monella enterica Serovar Typhimurium [25]; Vibrio algi-
nolyticus [26]; Citrobacter [27]; Vibrio fluvialis [28]; Vibrio
parahaemolyticus [29] and Shigella spp. [30].
Yeasts are ubiquitous in various aquatic environ-
ment, i.e. oceans, seas, estuaries, lakes and rivers [31].
Freshwater environments and marine waters receiving
organic loading supporting a high densities of C. albi-
cans which may be a health hazard [32]. Furthermore, a
higher incidence of vaginal infections caused by Can-
dida spp. among women who frequent beaches has
been reported [4]. The survival of C. albicans known as
pathogenic yeast in various water microcosms empha-
sizes the importance of assessing the quality of water
and providing a timely indication of bathing as well as
drinking water quality.
In this work, the viability of C. albicans in three wa-
ters microcosms (sea water, tap water and rain water)
at ambient temperature was investigated by using the
CFU counts method and the XTT reduction assay. The
obtained results for the different microcosms showed a
decrease in the CFU count and in the metabolic activity
reaching a low level of viability after 240 days. During
this starvation period, a gradual decrease in Candida
numbers was observed for all the water microcosms
with slight differences. Similar results has been re-
ported by Kashbur et al. [33].
Comparing the survival in the three microcosms, it
seems that C. albicans survive better in sea water after
240 days which may be due to the presence of high
concentration of some mineral elements such as Mg
2+
,
Ca
2+
, K
+
and Iron. These compounds may be used under
starvation conditions by C. albicans for their survival
during a long period of nutrient deprivation, confirm-
ing previous results suggesting their long term viability
in water [4, 34].
Our data showed also that viability increased slightly
at the first days and then decreased during starvation
(Figs. 1, 2 and 3). The Paired Samples Test revealed a
statistically significant difference between the control
(non starved cells) and the studied strain after one day
of incubation period.
The high oxidative activity of C. albicans during the
first days may be explained by the adaptation of cells in
their new environment rich in mineral elements sup-
porting earlier study which discribed that C. albicans
survive for extended period of time in aquatic envi-
ronment including marine and fresh water [32]. On the
other hand, Ahearn et al. [6] suggested that C. albicans
survive better in marine water than in fresh water. So,
it is clear that the two used methods gave almost the
same survival dynamics of C. albicans with a best survive
in sea water. The decrease of CFU was associated with
the diminution of the metabolic activity linked to the
decrease of cell viability. Our results proved the utility
of the use of metabolic assays method (XTT) as rapid
and high throughput analysis [20, 21, 35].
The molecular identification by PCR of the tested
strains has confirmed that the enumerated starved cells
in all water microcosms were C. albicans proving that

362 Kamel
Chaieb
et al.
Journal of Basic Microbiology 2011, 51, 357 – 363
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
C. albicans can survive in different water microcosms for
a long period which may be explained by their auto-
phagie strategy adopted under starvation condition [36].
During adaptation to new environments, C. albicans
need to recycle endogenous macromolecules in order to
provide nutrients [37]. In a recent study, Richards et al.
[38] reported that C. albicans can survive for six months
in water. Recently, a highest frequency of Candida was
reported in samples collected from the northern coast
of Saronicos gulf, in the Athens area, Greece [14] and in
drinking water [39]. In France, 50% of water samples
were contaminated by Candida [40] and in Finland,
these species occurred in 50% of chlorinated drinking
water samples [41].
In addition, Candida undergoes the transition from
blastospores to filaments in response to a wide variety
of conditions, such as body temperature (37 °C), or the
presence of some human hormones [42, 43]. One of the
strongest sets of filament-inducing conditions is the
combination of body temperature (37 °C) and serum
[44]. So, the CFU count seems not be influenced by fil-
amentous growth of Candida albicans cells in the studied
microcosms.
From the estimation of viability of C. albicans based
on CFU count versus the metabolic XTT assays, we can
conclude the persistence of such pathogenic yeast dur-
ing a long time in water. These results highlighted the
importance to include yeast in the microbiological
analysis of drinking water. Since these experiments
were carried out using pure culture suspensions, it is
difficult to estimate, the concentration and the survival
rate of these microorganisms in contaminated waters.
References
[1] Seneviratne, C.J., Jin, L., Samaranayake, L.P., 2008. Biofilm
lifestyle of Candida: a mini review. Oral. Dis., 14, 582–
590.
[2] Aronson, I.K., Soltani, K., 1976. Chronic mucocutaneous
candidosis: a review. Mycopathologia, 60, 17–25.
[3] Bonde, G.J., 1977. Bacterial indicators of water pollution.
Adv. Aquat. Microbiol., 1, 273–364.
[4] Buck, J.D., 1978. Comparison of an in situ and in vitro
survival of Candida albicans in seawater. Microb. Ecol., 4,
291–302.
[5] Hagler, A.N., Mendonca-Hagler, L.C., 1981. Yeasts from
marine and estuarine waters with different levels of pol-
lution in the state of Rio de Janeiro, Brazil. Appl. Environ.
Microbiol., 41, 173–178.
6] Ahearn, D.G., Roth, F.J., Meyers, J.S.P., 1968. Ecology and
characterization of yeasts from aquatic regions of South
Florida. Mar. Biol., 1, 291–308.
[7] Sherry, J.P., Kuchma, S.R., Dutka, B.J., 1979. The occur-
rence of Candida albicans in Lake Ontario bathing beaches.
Can. J. Microbiol., 25, 1036–1044.
[8] Hageskal, G., Knutsen, A.K., Gaustad, P., de Hoog, G.S.,
Skaar, I., 2006. Diversity and significance of mold species
in Norwegian drinking water. Appl. Environ. Microbiol.,
72, 7586–7593.
[9] Pereira, V.J., Basilio, M.C., Fernandes, D., Domingues, M.,
Paiva, J.M. et al., 2009. Occurrence of filamentous fungi
and yeasts in three different drinking water sources. Wa-
ter Res., 43, 3813–3819.
[10] Ikner, A., Shiozaki, K., 2005. Yeast signaling pathways in
the oxidative stress response. Mutat. Res., 569, 13–27.
[11] Albers, E., Larsson, C., Andlid, T., Walsh, M.C., Gustafs-
son, L., 2007. Effect of nutrient starvation on the cellular
composition and metabolic capacity of Saccharomyces cere-
visiae. Appl. Environ. Microbiol., 73, 4839–4848.
[12] Castellani, A., 1939. The viability of some pathogenic
fungi in sterile distilled water. J. Trop. Med. Hyg., 42,
225–226.
[13] Odds, F.C., 1991. Long-term laboratory preservation of
pathogenic yeasts in water. J. Med. Vet. Mycol., 29, 413–
415.
[14] Efstratiou, M.A., Tsirtsis, G., 2009. Do 2006/7/EC European
Union Bathing Water Standards exclude the risk of con-
tact with Salmonella or Candida albicans? Mar. Pollut. Bull.,
58, 1039–1044.
[15] Hartung de Capriles, C., Mata, S., Middelveen, M., 1989.
Preservation of fungi in water (Castellani): 20 years. My-
copathologia, 106, 73–79.
[16] Yamaguchi, H., Uchida, K., Nagino, K ., Matsunaga, T.,
2002. Usefulness of a colorimetric method for testing an-
tifungal drug susceptibilities of Aspergillus species to vori-
conazole. J. Infect. Chemother., 8, 374–377.
[17] Lattif, A.A., Mukherjee, P.K., Chandra, J., Swindell, K.,
Lockhart, S.R. et al., 2010. Characterization of biofilms
formed by Candida parapsilosis, C. metapsilosis, and C. or-
thopsilosis. Int. J. Med. Microbiol., 300, 265–270.
[18] McCluskey, C., Quinn, J.P., McGrath, J.W., 2005. An eva-
luation of three new-generation tetrazolium salts for the
measurement of respiratory activity in activated sludge
microorganisms. Microb. Ecol., 49, 379–387.
[19] Roehm, N.W., Rodgers, G.H., Hatfield, S.M., Glasebrook,
A.L., 1991. An improved colorimetric assay for cell proli-
feration and viability utilizing the tetrazolium salt XTT. J.
Immunol. Methods, 142, 257–265.
[20] Pettit, R.K., Weber, C.A., Kean, M.J., Hoffmann, H., Pettit,
G.R. et al., 2005. Microplate Alamar blue assay for Staphy-
lococcus epidermidis biofilm susceptibility testing. Antimic-
rob. Agents Chemother., 49, 2612–2617.
[21] Antachopoulos, C., Meletiadis, J., Roilides, E., Sein, T.,
Walsh, T.J., 2006. Rapid susceptibility testing of medically
important zygomycetes by XTT assay. J. Clin. Microbiol.,
44, 553–560.
[22] Chen, C.Y., Nace, G.W., Irwin, P.L., 2003. A 6 × 6 drop
plate method for simultaneous colony counting and MPN
enumeration of Campylobacter jejuni, Listeria monocytogenes,
and Escherichia coli. J. Microbiol. Methods, 55, 475–479.

Journal of Basic Microbiology 2011, 51, 357 – 363
Starvation survival of C. Albicans in water microcosmos
363
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
[23] Hoben, H.J., Somasegaran, P., 1982. Comparison of the
pour, spread, and drop plate methods for enumeration of
Rhizobium spp. in inoculants made from presterilized peat.
Appl. Environ. Microbiol., 44, 1246–1247.
[24] Turenne, C.Y., Witwicki, E., Hoban, D.J., Karlowsky, J.A.,
Kabani, A.M., 2000. Rapid identification of bacteria from
positive blood cultures by fluorescence-based PCR-single-
strand conformation polymorphism analysis of the 16S
rRNA gene. J. Clin. Microbiol., 38, 513–520.
[25] Ben Abdallah, F., Chaieb, K., Snoussi, M., Bakhrouf, A.,
Gaddour, K., 2007. Phenotypic variations and molecular
identification of Salmonella enterica serovar Typhimurium
cells under starvation in seawater. Curr. Microbiol., 55,
485–491.
[26] Ben Kahla-Nakbi, A., Besbes, A., Chaieb, K., Rouabhia, M.,
Bakhrouf, A., 2007. Survival of Vibrio alginolyticus in sea-
water and retention of virulence of its starved cells. Mar.
Environ. Res., 64, 469–478.
[27] Dhiaf, A., Bakhrouf, A., Witzel, K.P., 2008. Resuscitation
of eleven-year VBNC Citrobacter. J. Water Health, 6, 565–
568.
[28] Amel, B.K., Amine, B., Amina, B., 2008. Survival of Vibrio
fluvialis in seawater under starvation conditions. Microbi-
ol. Res., 163, 323–328.
[29] Abdallah, F.B., Kallel, H., Bakhrouf, A., 2009. Enzymatic,
outer membrane proteins and plasmid alterations of starv-
ed Vibrio parahaemolyticus and Vibrio alginolyticus cells in
seawater. Arch. Microbiol., 191, 493–500.
[30] Ellafi, A., Denden, I., Ben Abdallah, F., Souissi, I., Bakh-
rouf, A., 2009. Survival and adhesion ability of Shigella
spp. Strains after their incubation in seawater micro-
cosms. World J. Microbiol. Biotechnol., 25, 1161–1168.
[31] Fell, J., 2001. Collection and identification of marine
yeasts. In: Methods in Microbiology (Paul J., ed.). Aca-
demic Press: New York, 347–356.
[32] Valdes-Collazo, L., Schultz, A.J., Hazen, T.C., 1987. Survi-
val of Candida albicans in tropical marine and fresh waters.
Appl. Environ. Microbiol., 53, 1762–1767.
[33] Kashbur, I.M., Ayliffe, G.A., George, R.H., 1980. The survi-
val of Candida albicans in moist and dry environments. J.
Hosp. Infect., 1, 349–356.
[34] Ahearn, D.G., Meyers, S.P., Nichols, R.A., 1968. Extracellu-
lar proteinases of yeasts and yeastlike fungi. Appl. Micro-
biol., 16, 1370–1374.
[35] Ramage, G., Vande Walle, K., Wickes, B.L., Lopez-Ribot,
J.L., 2001. Standardized method for in vitro antifungal
susceptibility testing of Candida albicans biofilms. Anti-
microb. Agents Chemother., 45, 2475–2479.
[36] Mizushima, N., 2005. The pleiotropic role of autophagy:
from protein metabolism to bactericide. Cell Death Dif-
fer., 12, 1535–1541.
[37] Palmer, G.E., Kelly, M.N., Sturtevant, J.E., 2007. Autopha-
gy in the pathogen Candida albicans. Microbiology, 153,
51–58.
[38] Richards, D., Davies, J.K., Figdor, D., 2010. Starvation
survival and recovery in serum of Candida albicans com-
pared with Enterococcus faecalis. Oral Surg. Oral Med. Oral
Pathol. Oral Radiol. Endod., 110, 125–130.
[39] Burman, N.P., 1965. Taste and odor due to stagnation and
local warming in long lengths of piping. Proc. Soc. Water
Treat. Exam., 14, 125–131.
[40] Hinzelin, F., Block, J.C., 1985. Yeasts and filamentous
fungi in drinking water. Environ. Tech. Lett., 6, 101–106.
[41] Niemi, R.M., Knuth, S., Lundstrom, K., 1982. Actinomyce-
tes and fungi in surface waters and in potable water.
Appl. Environ. Microbiol., 43, 378–388.
[42] Brown, A.J., Gow, N.A., 1999. Regulatory networks cont-
rolling Candida albicans morphogenesis. Trends Microbiol.,
7, 333–338.
[43] Kinsman, O.S., Pitblado, K., Coulson, C.J., 1988. Effect of
mammalian steroid hormones and luteinizing hormone
on the germination of Candida albicans and implications
for vaginal candidosis. Mycoses, 31, 617–626.
[44] Kadosh, D., Johnson, A.D., 2005. Induction of the Candida
albicans filamentous growth program by relief of trans-
criptional repression: a genome-wide analysis. Mol. Biol.
Cell., 16, 2903–2912.
Wyszukiwarka
Podobne podstrony:
jobm 201000013
jobm 201000191
jobm 201000321
jobm 201000018
jobm 201000214
jobm 201000067
jobm 201000037
jobm 201000074
jobm 201000280
jobm 201000385
jobm 201000198
jobm 201000458
jobm 201000147
jobm 201000520
jobm 201000327
jobm 201000342
jobm 201000420
jobm 201000364
jobm 201000317
więcej podobnych podstron