
540
Journal of Basic Microbiology 2011, 51, 540 – 549
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
Research Paper
Caenorhabditis elegans as a model for
studying Cronobacter sakazakii ATCC BAA-894 pathogenesis
Bhagavathi Sundaram Sivamaruthi
1
, Abhijit Ganguli
2
, Mukesh Kumar
2
, Sheker Bhaviya
1
,
Shunmugiah Karutha Pandian
1
and Krishnaswamy Balamurugan
1
1
Department of Biotechnology, Alagappa University, Karaikudi, India
2
Department of Biotechnology & Environmental sciences, Thapar University, Patiala, Punjab, India
Cronobacter sakazakii is occasionally associated with food-borne illness seen in neonates and
infants with weakened immune system. It can cause meningitis, local necrotizing enterocolitis
and systemic bacteremia leading to infant mortality rates upto 33–80%. With the aim of
investigating whether C. sakazakii is also a pathogen of the model organism C. elegans, we have
performed killing assays and monitored the mortality of host fed with pathogen. C. elegans fed
with C. sakazakii die over the course of several days, as a consequence of an accumulation of
bacteria in the host intestine. Further, the rate of C. sakazakii mediated infection in C. elegans
depends on the accumulation of the bacterial load inside the host. C. sakazakii killed C. elegans
with an LT
50
(time for half to die) of 134 ± 2.8 h in liquid assay conditions, whereas the mor-
tality of C. elegans infected with C. sakazakii was less pronounced during solid assays. We found
that 24 h of C. sakazakii infection is enough to cause gametogenesis defects and increased cell
damage in intestinal tract of host. To monitor the immune regulations during C. sakazakii
infection in C. elegans at molecular level, total RNA was isolated and few candidate genes (lys-7,
clec-60 and clec-87) were kinetically analyzed by using the semi-quantitative RT-PCR. The level of
expression of lys-7, clec-60 and clec-87 mRNAs isolated from C. elegans infected with C. sakazakii
was significantly higher when compared to C. elegans exposed to E. coli OP50 control. This is the
first report in which physiological changes and an induction of host immunity mediated
antimicrobial genes by C. sakazakii are shown in C. elegans.
Abbreviations: NGM – Nematode growth medium; CFU – Colony Forming Unit, RT-PCR – Reverse
transcriptase-Polymerase chain reaction; PCR – Polymerase Chain Reaction
Keywords: Caenorhabditis elegans / Cronobacter sakazakii / Innate immune system
Received: September 20, 2010; accepted: December 23, 2010
DOI 10.1002/jobm.201000377
Introduction
*
Cronobacter sakazakii, a Gram-negative motile bacillus, is
an opportunistic pathogen that can cause infections in
blood stream and central nervous system [1, 2]. It has
been often associated with sporadic cases and is an
important causative agent of life threatening meningi-
tis (complicated by ventriculitis, brain abscess, cerebral
infraction and cyst formation), local necrotizing en-
Correspondence: Dr. K. Balamurugan, Department of Biotechnology,
Alagappa University, Karaikudi-630 003, India
E-mail: bsuryar@yahoo.com
Phone: 91-4565 225215
Fax: 91-4565 225202
terocolitis (NEC) and systemic bacteremia especially in
neonates and infants particularly in prematures with
weakened immune system [3–8]. C. sakazakii belongs to
the family of Enterobacteriaceae and was initially re-
ferred to as Enterobacter cloacae. An infant mortality rate
of 33–80% for C. sakazakii meningitis has been reported
with severe outcome of seizures, brain abscess hydro-
cephalus, developmental delay and death. Up to 20% of
the newborns developed serious neurological complica-
tions following infection [5, 6]. There are reports of
C. sakazakii infection in adults, which is not usually life
threatening [9]. One such report was vaginal infection
by C. sakazakii leading to vulvovaginitis with mucous

Journal of Basic Microbiology 2011, 51, 540 – 549
Caenorhabditis elegans, Cronobacter sakazakii and Innate
541
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
discharge [10]. Until recently, little is known about the
mechanisms involved in C. sakazakii pathogenesis. The
virulence study done by Mange et al. [6] observed no
relationship between the adhesive capacities in C. saka-
zakii and the eventual production of specific fimbriae
during the establishment of pathogenic infection.
Townsend et al. [22] demonstrated the influence of en-
dotoxin on increased translocation of intestinal bacte-
ria and C. sakazakii in the neonatal rat. These studies
further demonstrated the ability of C. sakazakii to in-
vade brain capillary endothelial cells and in human
macrophages. In mammalian cells, the organism can
attach to intestinal cells and survive internally in
macrophages [4]. However, the specific bacterial adhe-
sins and host cell receptors involved in these processes
are largely unknown. Some strains of C. sakazakii pro-
duce capsular material, and how this material contrib-
utes to macrophage evasion remains to be determined
[4]. Furthermore, these capsules may also provide pro-
tection for the organism, facilitating its survival in
dessicated environments [4]. A recent study by Kim et al.
revealed that OmpX and OmpA are involved in baso-
lateral invasion by C. sakazakii [11].
Caenorhabditis
elegans is a free-living, non-parasitic ne-
matode, which is widely used as a model to study bac-
terial pathogenesis because of the fact that the genetic
and molecular tools used for its manipulation are well
developed. Furthermore, the simple growth conditions,
rapid generation time with an invariant cell lineage and
importantly, the known genome sequences made this
animal model to study complicating processes at both
the physiological and cellular levels [12, 13]. In this stu-
dy, pathogenicity of C. sakazakii using the model organ-
ism, C. elegans was investigated. C. elegans usually grinds
the bacteria and extracts the nutrients from them. The
phenomenon of bacterial cell escaping the pharyngeal
pumping of C. elegans indicates the invasion capability
of the bacteria [14]. Using both solid and liquid culture
based killing assays and microscopic observations the
host responses at the physiological levels were studied.
Analysis of candidate antimicrobial genes at their
mRNA levels revealed a substantial relation between C.
sakazakii-mediated innate immune gene responses in
the host system.
Materials and methods
Bacterial strains, nematode and reagents
The wild type strain Cronobacter sakazakii ATCC BAA-894
was grown on tryptic soy agar (TSA) (Himedia, Mumbai,
India). Escherichia coli OP50 was, provided by the CGC
(Caenorhabditis Genetic Centre), grown on Luria Ber-
tani (Himedia) agar plates. All bacterial strains were
grown overnight at 37 °C in Luria broth (LB). C. elegans
strain N2 Bristol was maintained on Nematode Growth
Medium [NGM, minimal medium containing NaCl,
agar, peptone, cholesterol, CaCl
2
, MgSO
4
, and potassium
phosphate] [12] containing a lawn of E. coli OP50 (food
source) at 20 °C. All experiments were carried out using
age-synchronized L4 stage animals. Synchronous popu-
lations were acquired by bleaching the gravid adults.
The bleached eggs were allowed to hatch and develop
into L4 at 20 °C. Synchronized L4 stage worms were
collected by using M9 solution and were washed several
times and used for different assays.
Liquid killing assay on C. elegans
Unless otherwise specified, most of the assays were
performed under liquid conditions. Briefly, 2 ml of LB
was inoculated with a single colony of the appropriate
bacterial strain and grown at 37 °C for 3 h. 100 μl of 3 h
culture of C. sakazakii or E. coli OP50 (control) was added
to 400 μl of M9 buffer containing known number of
worms (~20 L4 stage hermaphrodites) in each well of
micro titer well plate. Negative control is the well con-
taining M9 buffer and nematodes without any bacterial
culture. The worms were monitored for every 1 h inter-
val to note the exact time required for its death on
continuous exposure to pathogen. Worms were consid-
ered dead when they did not respond to touch with a
platinum wire pick and without any pharyngeal
movement for several hours. Each experimental condi-
tion was tested in triplicates.
Solid killing assay with C. elegans
For the assays performed on solid NGM-agar plates,
bacterial lawns were grown for C. elegans killing assays
as follows: Individual bacterial colonies were inoculated
into 2 ml of LB and grown at 37 °C for 3 h; 100 μl of the
culture of 0.5 OD was spread on 35-mm Petriplates
containing NGM. The plates were incubated at room
temperature for overnight. Approximately, twenty num-
bers of age-synchronized L4 stage hermaphrodites were
transferred from a lawn of E. coli OP50 to a lawn of the
bacterium to be tested, incubated at 20 °C, and exam-
ined at 24 h intervals with a dissecting microscope for
viability. Worms were considered dead upon failure to
respond to touch. Each experimental condition was
tested in triplicates.
Short-time exposure assays on C. elegans
The age-synchronized L4 stage hermaphrodites were
exposed to C. sakazakii liquid culture for different time

542 B.
S.
Sivamaruthi
et al.
Journal of Basic Microbiology 2011, 51, 540 – 549
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
intervals and washed with M9 buffer to remove the
bacterial cells which is adhered on the surface of the
worm and transferred to NGM plates (seeded/unseeded
with E. coli OP50) to monitor their behavioral changes
and survival.
Bacterial accumulation assay
To correlate C. sakazakii killing of C. elegans with bacte-
rial load inside the exposed worm’s gut, bacterial ac-
cumulation assay was performed. To examine bacterial
accumulation, L4 stage hermaphrodites were trans-
ferred to E. coli OP50 or C. sakazakii bacterial culture (as
like liquid killing assay) and were incubated at 20 °C.
After every 24 h of exposures, worms were serially
washed with M9 buffer containing 1 mM sodium azide
to inhibit expulsion of bacteria from their intestine.
The nematodes were treated with antibiotic (1 mg/ml
gentamicin) to wash the pathogen adhered on the outer
surface, lysed in phosphate-buffered saline (PBS) con-
taining 400 mg of 1.0 mm silicon carbide particles
(Himedia) and mechanically disrupted using a pestle.
The worm lysates were diluted and plated on TSA
plates. The plates were incubated overnight at 37 °C.
Colonies were quantified and used to calculate the
number of bacteria per nematode.
Sytox staining
By measuring the uptake of Sytox Green nucleic acid
stain by the dead cells, the dead versus live C. elegans
cells were determined. To study the cell damage, nema-
todes were exposed to E. coli OP50 or C. sakazakii bacte-
rial culture at 20 °C for 24 h. After washing with M9
buffer, the worms were treated with 1 μM of Sytox
stain for 5 min and remove excess dye on the surface of
worm by washing with M9 buffer. The nematodes were
then placed on a pad of 2% agarose in a 5 μl drop of
30 mM NaN3 in M9 medium and observed under in-
verted fluorescent microscope (Nikon Eclipse Ti-S, Ja-
pan).
Pharyngeal pumping assay
To determine the pumping rate, worms were placed on
NGM plates seeded with E. coli OP50 (control) and
C. sakazakii. The pharyngeal pumping was observed
using a stereomicroscope (Nikon SMZ1000, Japan) for
thirty consecutive seconds.
Chemotaxis assay
The cultures (0.2 OD) of C. sakazakii and E. coli OP50 were
spotted at a distance of 3 cm from the centre of NGM
plates (90 mm) and denoted as zone A and zone B, re-
spectively. Twenty-five wild type C. elegans were thor-
oughly washed from E. coli OP50 lawn and placed at the
center of the NGM plate. The number of worms in zone
A and zone B were counted every 4 h. Similarly in con-
trol plates, both zone A and zone B were spotted with
OP50 or C. sakazakii, respectively.
Microscopic analysis
Nematodes were exposed to C. sakazakii or E. coli OP50
for various time intervals and then placed on a pad of
2% agarose (20 mg of Agarose was dissolved in 1 ml of
distilled water by gentle heating and a drop of boiled
agarose placed on a glass slide and flat surface made by
placing another glass slide on the drop. After solidifica-
tion, the top glass slide were removed gently) in a 5 μl
drop of 30 mM NaN
3
in M9 medium. The worms were
examined for their physiological changes under the
microscope and Confocal Laser Scanning Microscope
(Carl Zeiss, Germany).
Preparation of worm total RNA and semi-quantitative
RT-PCR
Synchronized populations of wild-type L4 stage worms
were generated at 20 °C on the standard food source
from eggs. Worms were collected from E. coli OP50
lawns in M9 buffer at room temperature and washed
twice with M9 buffer. Nematodes were exposed to
C. sakazakii or E. coli OP50 for different time intervals at
Table 1. Primer sequences used for the semi-quantitative RT-PCR analysis.
Target
Primer
Sequence
Amplicon size (in ~bp)
Forward 5′-ATCGTCCTCGACTCTGGAGATG-3′
act-2
Reverse 5′-TCACGTCCAGCCAAGTCAAG-3′
101
Forward 5′-TTGCAGTACTCTGCCATTCG-3′
lys-7
Reverse 5′-GCACAATAACCCGCTTGTTT-3′
199
Forward 5′-TGTCTGCATTCTTCCAGTCG-3′
clec-60
Reverse 5′-CCCATACCCAGACACCTTTG-3′
197
Forward 5′-AATTCGTGTTCAAGCCAAGG-3′
clec-87
Reverse 5′-AGCCAGTTGATTTTGGTTGG-3′
132
Journal of Basic Microbiology 2011, 51, 540 – 549
Caenorhabditis elegans, Cronobacter sakazakii and Innate
543
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
20 °C. The worms were washed and treated with TRIzol
reagent. The total RNA from whole worms was ex-
tracted according to the protocol of R. D. Burdine and
M. J. Stern (http://www.wormbase.org; WormBase re-
lease WS188, WBPaper00015272). Total RNA isolated
from the infected L4 stage C. elegans and their respective
control was reverse-transcribed using oligodT primer
and Superscript III as per the manufactures instruc-
tions (Invitrogen). After first strand synthesis, PCR were
carried out to analyze the expression pattern of candi-
date antimicrobial genes using gene specific primers
(Table 1).
Statistical analysis
The data represented as mean survival time ± SD in
hours for N2 worms infected during L4 stage at 20 °C.
Post Hoc Tests was performed to evaluate the signifi-
cant differences (at 0.05 level) between the survival
curve of C. elegans exposed to the pathogen C. sakazakii
and the control E. coli OP50.
Results
C. sakazakii killing of C. elegans correlates
with bacterial accumulation in the intestine
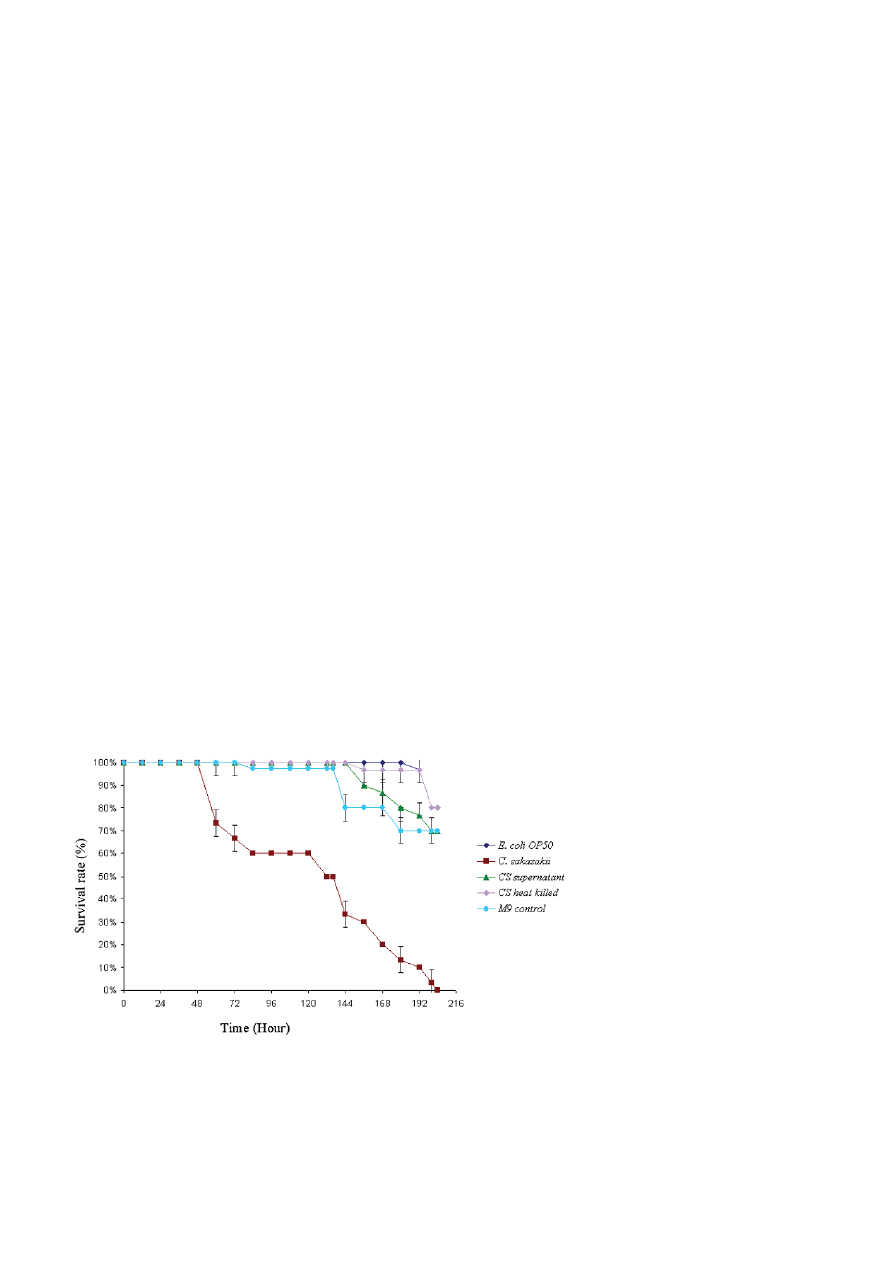
C. sakazakii was capable of killing C. elegans. As shown in
Fig. 1 wild type N2 nematodes died rapidly when fed on
C. sakazakii than on E. coli OP50 (the usual food source
for propagating C. elegans in the laboratory) under liq-
uid condition. The LT
50
for C. sakazakii was calculated in
three independent experiments (LT
50
of 134 ± 2.8 h) and
determined to be significantly shorter when compared
to the nematodes feeding on E. coli OP50 (Fig. 1). In the
negative control (bacteria-free media), the mortality of
C. elegans occurred by internal hatching of progeny due
to lack of food source
.
In addition, we examined
whether C. sakazakii killing of C. elegans correlated with
bacterial accumulation in the intestine (Fig. 2A). The
profile of bacterial accumulation in the gut was exam-
ined by scoring the number of live bacteria in the gut
and confirmed by direct observation under the micro-
scope (Fig. 2B and 2C) [15]. As shown in Fig. 2, the ac-
cumulation of C. sakazakii in the nematodes was in-
creased with time during the course of infections.
Characterization of C. sakazakii mediated killing
of C. elegans
According to Mylonakis and Aballay, short exposures to
S. enterica, as little as 5 h resulted in a persistent and
lethal infection of C. elegans that correlated with bacte-
rial replication in the intestinal lumen [16]. To deter-
mine whether C. sakazakii was capable of persistently
colonizing C. elegans intestine, we exposed C. elegans to
C. sakazakii for 2–24 h and then transferred the worms
to NGM plate (seeded with E. coli OP50) and compared
their survival rate to that of worms either continuously
exposed to E. coli OP50 or C. sakazakii. C. elegans exposed
to C. sakazakii for 2–12 h exhibited normal life span,
like those fed with E. coli OP50 for the same duration of
the assay (Table 2). However, worms exposed to C. saka-
zakii for 24 h were paralyzed, exhibited defects in game-
Figure 1. Survival of C. elegans fed on pathogenic C. sakazakii strain (
䊏
), cell free C. sakazakii supernatant (
䉱
), heat killed C. sakazakii
(
䉬
) E. coli OP50 control (
䉬
) and M9 control (
䊉
) in liquid condition. The differences between the life span of C. elegans exposed to E. coli
OP50 (control) (
䉬
) and worms exposed to C. sakazakii (
䊏
) are significant (p < 0.001). The differences between the survival of C. elegans
exposed to E. coli OP50 control and C. elegans exposed to cell free C. sakazakii supernatant (
䉱
) or heat killed C. sakazakii (
䉬
) are not
significant. P-values were generated by ANOVA (Dunnett’s T3 post hoc test). P < 0.05 was considered to be significant.

544 B.
S.
Sivamaruthi
et al.
Journal of Basic Microbiology 2011, 51, 540 – 549
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
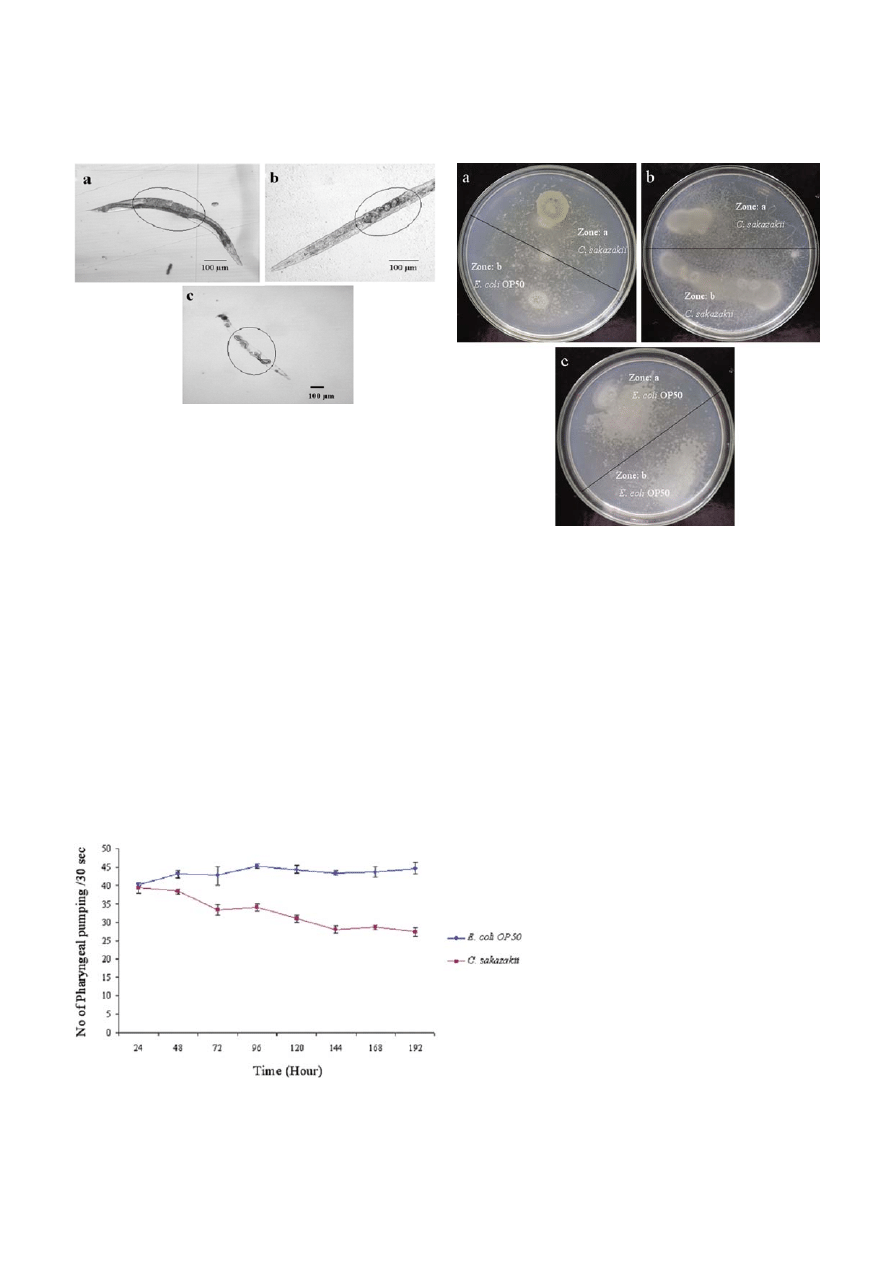
Figure 2. Bacterial accumulation in C. elegans: A) The representative image showing an increased colonization of C. sakazakii at the
intestinal region of the infected animal [at various time exposures as 48 (a), 96 (b), 144 (c), and 192 h (d), respectively] when compared to
the worms fed with E. coli OP50 (f). Accumulation of pathogen inside the worms was indicated as circle and arrows. The status of eggs in
control and infected worms were also indicated with arrows (e and f, respectively). B) The bacterial load (Colony Forming Unit; CFU) in
C. elegans exposed to C. sakazakii (
䊏
) was analyzed as described in ‘Materials and methods’. C) Microscopic observations showing the
increased colonization (circled) of C. sakazakii in the post pharyngeal region of C. elegans [at various time exposures as 144 (b), and 192 h
(d), respectively] when compared to the control (a and c).
togenesis (Fig. 3) and died at 120 h even in the presence
of food source, suggesting that, C. sakazakii infection in
C. elegans requires at-least 24 h and 120 h of exposures
for paralysis and mortality respectively.
In another experiment, C. elegans either exposed to
E. coli OP50 or C. sakazakii for 24 h were then transferred
back to E. coli OP50 and monitored for the internalized
bacteria. C. elegans were removed after every 24 h, col-
lected in a tube and mechanically disrupted to release
the internalized bacteria, which were quantified using
a plating assay. The numbers of internalized C. sakazakii
increased over time (after 24 h) (Fig. 2B), indicating that
C. sakazakii could persistently multiply and colonize the
intestine of the exposed C. elegans. We examined whe-
ther C. sakazakii might kill C. elegans using a mechanism
that involves diffusible toxins. Hence, we examined
whether live C. sakazakii was required for C. elegans mor-
tality by feeding nematodes on either live or heat-killed
C. sakazakii. Though the exposed C. elegans exhibited a
significantly reduced life-span against live C. sakazakii,
there was no related decrease in the nematode life-span
by the heat-killed C. sakazakii cultures (Fig. 1). In an-
other experiment, a group of L4 stage worms were ex-
posed to culture free supernatant of C. sakazakii and
analyzed for the changes in their life-span. As shown in
Fig. 1, we did not observe decreased life-span of the
nematodes treated with culture free supernatant of
C. sakazakii, suggesting that C. sakazakii appeared to not
kill C. elegans by producing any externally secreted
toxin(s). The differences between C. elegans exposed to
C. sakazakii and E. coli OP50 (p < 0.001) are significant.
The differences between the survival of C. elegans ex-
Table 2. Status of pathogen exposed to C. elegans is in different conditions (seeded and unseeded NGM plates).
Time of Exposure (hour)
On Seeded NGM
(with
E. coli
OP50)
On unseeded NGM
(without laboratory food source)
2
Active & produced progeny on the third day
Active & produced progeny after five days
4
Active & produced progeny on the third day
Active & produced progeny after five days
8
Active & produced progeny
Active & produced progeny
12
Active & produced progeny
Active & produced progeny on the third day
24
Animals were paralyzed and died without
progeny after five days
Animals were paralyzed and dead after third day
Journal of Basic Microbiology 2011, 51, 540 – 549
Caenorhabditis elegans, Cronobacter sakazakii and Innate
545
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
Figure 3. Improper development of eggs (indicated as circle) in
C. sakazakii infected nematode (a), healthy development of eggs in
E. coli OP50 fed nematode (b) and internal hatching in M9 control in
liquid infection assay.
posed to E. coli OP50 control and the cell free C. sakazakii
supernatant or heat killed C. sakazakii exposed to C. ele-
gans are not significant.
The study by Shtonda and Avery confirmed that
some bacterial species do not support the growth of the
worm adequately [17] and hence chemotaxis assay was
performed to analyze the olfactory response of C. ele-
gans against C. sakazakii. The result of this assay con-
firmed that C. elegans didn’t show pathogen avoidance
behaviour against C. sakazakii (Fig. 4). However, the data
obtained from the pharyngeal pumping assay indicated
that C. sakazakii have an impact on pharyngeal pumping
rate of infected C. elegans (Fig. 5).
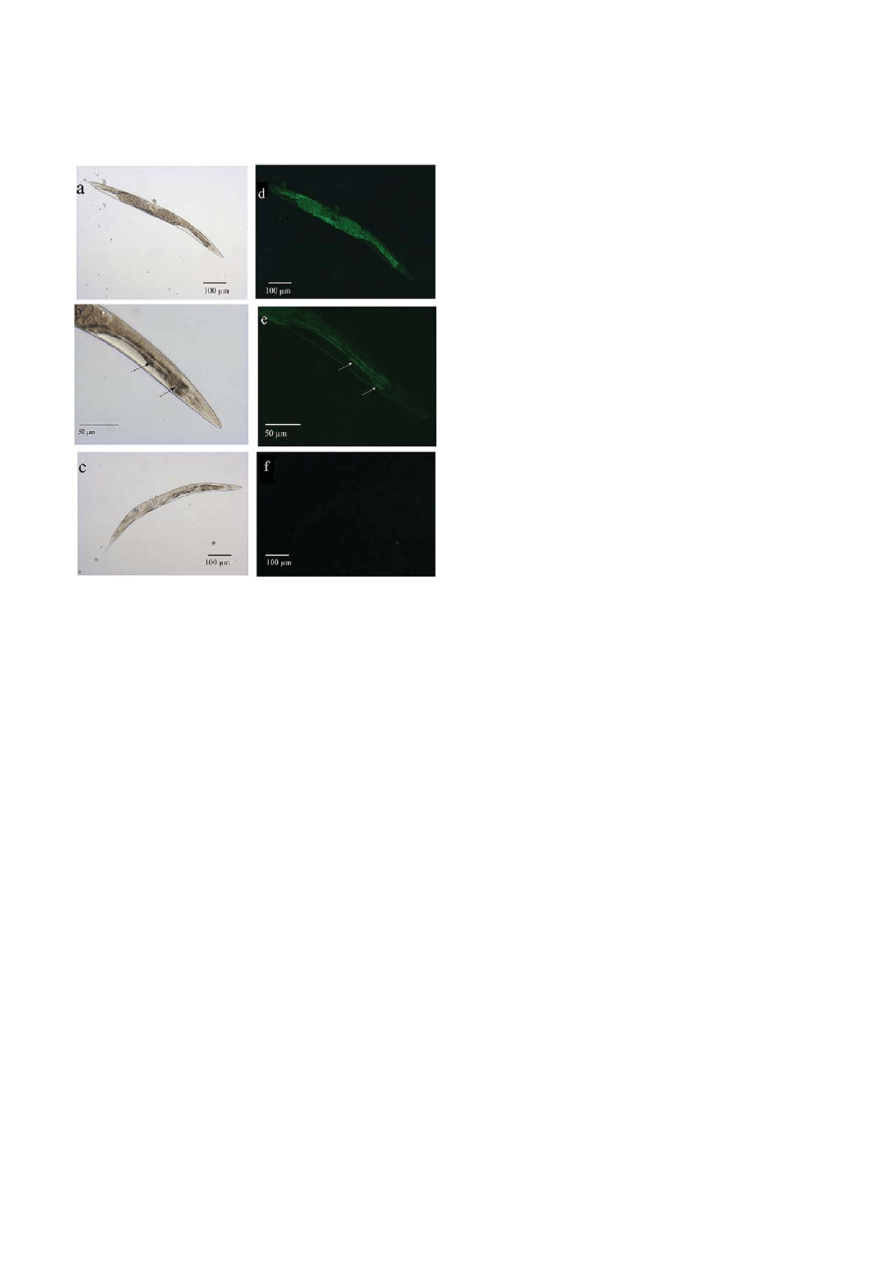
In order to observe the possibility of any cell damage
in C. elegans by the exposure with C. sakazakii, we ex-
posed C. elegans to C. sakazakii for 24 h and then stained
with nucleic acid stain Sytox to observe the damaged or
Figure 4. Images showing Chemotaxis behaviour of C. elegans
against C. sakazakii. a) Worms in presence of food source, E. coli
OP50 (marked as zone ‘b’) and pathogen source, C. sakazakii
(marked as zone ‘a’). The worms moved freely towards both the
zones. b). Worms crawling in the spots of C. sakazakii (at both zone
A and zone B). The worms moved towards both the zones. c)
Worms in presence of food source, E. coli OP50 (marked as zone
‘a’ and zone ‘b’).
apoptotic cells within the nematode body (Fig. 6). The
results indicated that the cells of intestinal lumen and
other parts of nematode were deeply damaged by
pathogenic infection when compared to E. coli OP50
treated C. elegans, suggesting that C. sakazakii could kill
C. elegans by inducing increased level of apoptosis and
bacterial accumulation.
Figure 5. Pharyngeal pumping rate (number of flings per 30 seconds) of control (
䉬
) and C. sakazakii infected C. elegans (
䊏
) during the
exposures.
546 B.
S.
Sivamaruthi
et al.
Journal of Basic Microbiology 2011, 51, 540 – 549
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
Figure 6. Sytox nucleic acid staining pattern of C. elegans infected
with C. sakazakii. Representative C. elegans exhibited a drastic
damage in intestinal track (arrow indication) and other parts of the
worm when fed with C. sakazakii (a, b, d and e) compared to
C. elegans fed with E. coli OP50 (c and f).
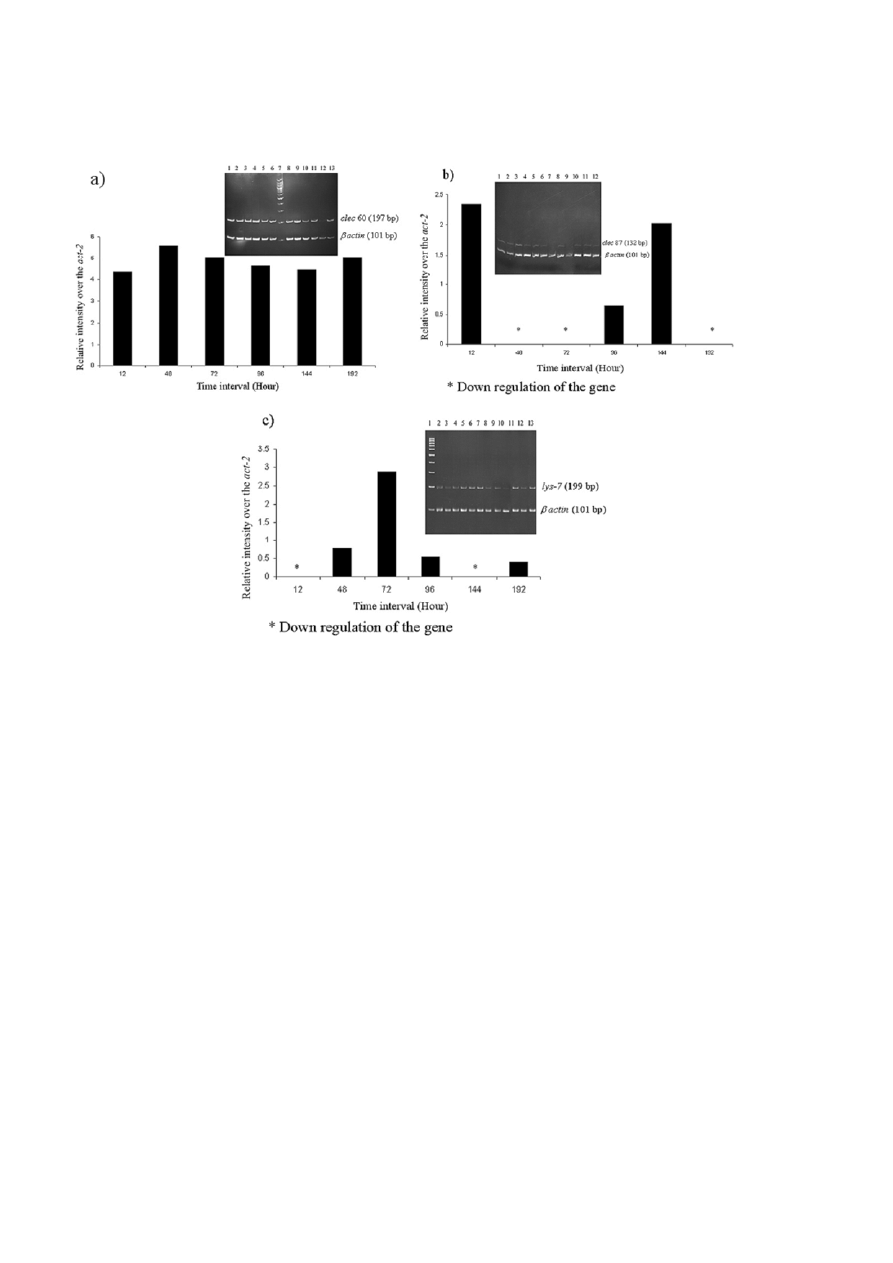
Role of candidate immune genes (lys-7, clec-60
and clec-87) during C. sakazakii infections
There are many putative antimicrobial effectors en-
coded in C. elegans genome and were shown to be in-
duced by infections [18]. We assessed whether C. elegans
respond to C. sakazakii infection with a quantifiable
defense response. Candidate genes (lys-7, clec-60 and
clec 87) known to be associated with C. elegans innate
immune response to pathogenic infections was used as
indicators to assess the transcriptional profile of worms
exposed to C. sakazakii. The Semi-quantitative RT-PCR
analyses clearly revealed the level of expression of lys-7,
clec-60 and clec-87 mRNAs during the pathogenesis of
C. sakazakii on C. elegans. The data revealed that the ex-
pressions of these candidate genes are notably higher
than their respective control(s). Particularly, the mRNA
level of lys-7, a known antimicrobial effector gene ap-
peared to be upregulated during the initial phases of
infection (till 72 h) and was down regulated during the
latter hours of infections indicating the surrendering of
host innate immune system during C. sakazakii patho-
genesis. However, the expression levels of both clec-60
and clec-87 appeared to be not-regulated during the
pathogenesis of C. sakazakii, thus emphasizing their
roles as pathogen recognizing factors for the externally
interacting pathogens (Fig. 7a–c). The relative intensity
of bands on the gel was measured using ImageJ sup-
ported these results.
Discussion
C. elegans are susceptible to a number of bacterial pa-
thogens, which kill the nematodes using a variety of
mechanisms, such as Pseudomonas aeruginosa PA14 pro-
duces phenazines, which are toxic to the nematode [19,
20] and S. enterica, establish a persistent infection
within the gut of the nematode [15, 21]. The present
study aims to investigate both physiology and the mo-
lecular responses of a host system (C. elegans) exposed
with C. sakazakii. The virulence factors of C. sakazakii
leading to pathogenesis in infants causing meningitis,
necrotizing enterocolitis, sepsis still have to be deter-
mined, although few studies related to the invasiveness
and adhesion done by Mange et al. [6] and Townsend
et al. [2]. However, the actual mechanism behind the
infection has not been explored. To evaluate whether
C. elegans model will be useful for studying and identify-
ing C. sakazakii virulence factors relevant to infant dis-
ease, we initially, analyzed the interaction of C. saka-
zakii with C. elegans at physiological and transcriptional
level. We observed that C. sakazakii killed C. elegans with
an LT
50
of 134 ± 2.8 h and short time of pathogenic
exposure, minimum of 24 h, is sufficient to infect and
kill the nematode. The differences between the mortal-
ity of C. elegans exposed to C. sakazakii and control E. coli
OP50 (p < 0.001) are significant. As shown in Fig. 5,
C. sakazakii infection has an impact on pharyngeal
pumping rate of C. elegans. Pharyngeal pumping rate of
C. elegans infected with C. sakazakii have been reduced in
a time dependent manner i.e, when the duration of
worm exposure to pathogen increased, the pumping
rate was reduced gradually. Progress in pharyngeal
damage was also observed, compared to control, during
the in-fection course, which was imaged microscopi-
cally (Fig. 2C).
C.
elegans infected with C. sakazakii produced drasti-
cally fewer progenies during liquid assay compared to
worms fed on E. coli OP50 (Fig. 3). Though, C. elegans
exposed to pathogen reproduced to some extent on full-
lawn (Solid NGM Plates) of C. sakazakii, the number of
individuals in successive generations declined by more
than 30% in solid assay (data not shown). As shown in
Fig. 3, C. sakazakii infection could contribute to the de-
creased numbers of healthy egg production (28 ± 4)
Journal of Basic Microbiology 2011, 51, 540 – 549
Caenorhabditis elegans, Cronobacter sakazakii and Innate
547
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
Figure 7. Transcriptional profiles of clec-60, clec-87 and lys-7 during C. sakazakii exposures. The semi-quantitative RT-PCR of the candi-
date genes was performed as described in Materials and Methods. The PCR products were analyzed during the linear range of ampli-
fications on a 12% PAGE. The relative intensities were compared with internally amplified house keeping control gene, act-2. The inlet
figure showed the amplicons on a 12% PAGE. a) The level of expression of clec-60 mRNA in C. sakazakii infected C. elegans of different
time intervals as 12, 48, 72, 96 and 144 h, (Lane: 1, 3, 5, 8, 10 and 12) respectively, and C. elegans exposed to E. coli OP50 control for 12,
48, 72, 96, 144 h, (Lane: 2, 4, 6, 9, 11 and 13) respectively. Lane 7 shows the 100 bp DNA ladder. b) The level of expression of clec-87
mRNA in C. sakazakii infected C. elegans of different time intervals as 12, 48, 72, 96, 144 h, (Lane: 1, 3, 5, 7, 9 and 11) respectively and
C. elegans exposed to E. coli OP50 control for 12, 48, 72, 96, 144 h, (Lane: 2, 4, 6, 8, 10 and 12) respectively. c) The level of expression of
lys-7 mRNA in C. sakazakii infected C. elegans of different time intervals as 12, 48, 72, 96, 144 h, (Lane: 2, 4, 6, 8, 10 and 12) respectively
and C. elegans exposed to E. coli OP50 control for 12, 48, 72, 96, 144 h, (Lane: 3, 5, 7, 9, 11 and 13), respectively. Lane 1 shows the
100 bp DNA ladder. *denotes the down regulation of the respective mRNAs over the house keeping gene.
compare to control (283 ± 6). Townsend et al. showed
that C. sakazakii are able to penetrate rat brain endothe-
lial cells and to survive inside macrophages [22]. Kim
and Loessner [9] demonstrated that C. sakazakii are able
to invade human intestinal epithelial cells. C. sakazakii
has an invasion mechanism different from those em-
ployed by L. monocytogenes and Salmonella serovar Typhi-
murium [9]. In this study, we demonstrated that C. saka-
zakii infected C. elegans for 24 h displayed CFU of 3.25 ×
10
4
per worm and the gradual multiplication of C. saka-
zakii was observed during course of infection as shown
in Fig. 2B (CFU expressed as log value of CFU ml
–1
) sug-
gesting that the pathogenicity exhibited by C. sakazakii
was appeared to be mainly due to the bacterial accu-
mulation and persistent infection inside the host sys-
tem.
It is known that the model organism, C. elegans en-
counters many strains of bacteria in its natural soil
environment. The ability to find good food sources over
potential pathogens is a significant advantage in using
C. elegans for physiological studies pertaining to host-
pathogen interactions. It uses a sophisticated chemo-
sensory system to identify food and olfactory learning
as a mechanism to avoid pathogens (23) Although, re-
cent studies have suggested that this conditioning be-
havior was analogous to mammalian taste aversion, a
continuous exposure to pathogen might make C. elegans
under stress to avoid pathogenic bacteria for long dura-
tion. To avoid such factors in understanding the mini-
mum time required for a pathogen to infect a host, we
have performed short-time exposure assays under liq-
uid conditions. The worms exposed to C. sakazakii for

548 B.
S.
Sivamaruthi
et al.
Journal of Basic Microbiology 2011, 51, 540 – 549
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
24 h in the presence of food source, exhibited defects in
gametogenesis, paralyzed, and died after 120 h (Fig. 3),
suggesting that C. sakazakii infection in C. elegans re-
quired at-least 24 h of exposures that resulted in worm
mortality after 120 h even in the presence of E. coli
OP50. Chemotaxis assay confirmed that the pathogen
avoidance behaviour was not exhibited by C. elegans
against C. sakazakii. In addition, the intake of C. sakazakii
into the exposed worms was not as significant as been
observed with liquid assays. The solid assay on NGM
plates did not exhibit significant impact on the life
span of exposed C. elegans (data not shown). The number
of worms in both spots of E. coli OP50 and C. sakazakii
were found to be even after 24 h, indicating that olfac-
tory system of C. elegans doesn’t response well against C.
sakazakii (Fig. 4). These results also indicate that C. ele-
gans intakes the pathogen C. sakazakii and the mortality
of worms are mainly due to the pathogenic impact of
C. sakazakii and not due to starvation.
As shown in Fig. 6, the nematode infected with
C. sakazakii exhibited drastic damage in intestinal region
and other parts of the worms when compared to
C. elegans fed with E. coli OP50, suggesting that C. saka-
zakii has induced apoptosis (cell death was confirmed by
Sytox staining) in host system appeared as the major
event during the pathogenesis of C. sakazakii.
To evaluate the transcriptional response of C. elegans
infected with C. sakazakii at the immune level, the
mRNAs of two C-type lectin genes (clec-60, clec-87) and a
lysozyme gene (lys-7) was studied kinetically. The tran-
scriptional response of candidate genes clec-60, clec-87
and lys-7 by semi-quantitative RT-PCR assessments were
taken primarily as an indication of C. elegans exposed
with C. sakazakii and was considered as a result of
pathogen induced host innate immune response(s).
In conclusion, we have confirmed that C. sakazakii
causes infection and leads to the mortality of C. elegans
by an active process that required live infection, which
correlated with the cell damage, bacterial accumulation
and persistent infection in the intestine. We trust that
C. elegans – C. sakazakii ATCC BAA-894 pathogenesis sys-
tem may be useful for studying interactions between
Cronobacter and invertebrates, besides the data gathered
from the present study will be useful in developing
intervention strategies to control C. sakazakii infection.
References
[1] Iversen, C., Lehner, A., Mullane, N., Bidlas, E. et al., 2007.
The taxonomy of Enterobacter sakazakii: proposal of a new
genus Cronobacter gen. nov. and descriptions of Cronobacter
sakazakii comb. nov. Cronobacter sakazakii subsp. sakazakii,
comb. nov., Cronobacter sakazakii subsp. malonaticus subsp.
nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii
sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter
genomospecies I. BMC Evol. Biol., 7, 64.
[2] Dumen, E., 2010. Cronobacter sakazakii (Enterobacter saka-
zakii): only an infant problem? Kafkas Univ Vet Fak Derg.,
16, S171–S178.
[3] Bowen, A.B., Braden, C.R., 2006. Invasive Enterobacter
sakazakii disease in infants. Emerg. Infect. Dis., 12, 1185–
1189.
[4] Drudy, D., Quinn, N.R., Wall, P.G., Fanning, S., 2006.
Enterobacter sakazakii: an emergent pathogen in powdered
infant formula. Clin. Infect. Dis., 42, 996–1002.
[5] Kim, J.B., Cho, S.H., Park, Y.B., Lee, J.B. et al., 2008. Sur-
veillance of stool samples for the presence of Enterobacter
sakazakii among Korean people. Yonser. Med. J., 49, 1017–
1022.
[6] Mange, J.P., Stephan, R., Borel, N., Wild, P. et al., 2006.
Adhesive properties of Enterobacter sakazakii to human epi-
thelial and brain microvascular endothelial cells. BMC
Microbiol., 6, 1–10.
[7] Mullane, N., Gaora, P.O., Nally, J.E., Iversen, C. et al.,
2008. Molecular analysis of the Enterobacter sakazakii O –
antigen gene locus. Appl. Environ. Microb., 74, 3783–
3794.
[8] Erickson, M.C., Kornacki, J.L., 2002. Enterobacter sakazakii:
An emerging food pathogen.
Acedido. em. Fev.,
25, 2008.
[9] Kim, K.P., Loessner, M.J., 2008. Enterobacter sakazakii inva-
sion in human intestinal Caco – 2 cells requires the host
cell cytoskeleton and is enhanced by disruption of tight
junction. Infect. Immun., 76, 562–570.
[10] Ongradi, J., 2002. Vaginal infection by Enterobacter sakaza-
kii. Sex. Transm. Infect., 78, 467–468.
[11] Kim, K., Kim, K.P., Choi, J., Lim, J.A. et al., 2010. Outer
membrane proteins A (OmpA) and X (OmpX) are essential
for basolateral invasion of Cronobacter sakazakii. Appl. En-
viron. Microbiol., 76, 5188–5298.
[12] Brenner, S., 1974. The genetics of Caenorhabditis elegans.
Genetics, 77, 71–94.
[13] Darby, C., 2005. Interactions with microbial pathogens.
WormBook. pp. 1–15.
[14] Tenor, J.L., Aballay, A., 2008. A conserved Toll-like recep-
tor is required for Caenorhabditis elegans innate immunity.
EMBO Rep., 9, 103–109.
[15] Aballay, A., Yorgey, P., Ausubel, F.M., 2000. Salmonella
typhimurium proliferates and establishes a persistent in-
fection in the intestine of Caenorhabditis elegans. Curr. Biol.,
10, 1539–1542.
[16] Mylonakis, E., Aballay, A., 2005. Worms and flies as gene-
tically tractable animal models to study host–pathogen
interactions. Infect. Immun., 73, 3833–3841.
[17] Shtonda, B.B., Avery L., 2005. Dietary choice behavior in
Caenorhabditis elegans. J. Exp. Biol., 209, 89–102.
[18] O’Rourke, D., Baban, D., Demidova, M., Mott, R. et al.,
2006. Genomic clusters, putative pathogen recognition
molecules, and antimicrobial genes are induced by infec-
tion of C. elegans with M. nematophilum. Genome Res., 16,
1005–1016.

Journal of Basic Microbiology 2011, 51, 540 – 549
Caenorhabditis elegans, Cronobacter sakazakii and Innate
549
© 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
www.jbm-journal.com
[19] Miklos, S.M., Tan, M.W., Rahme, L.G., Ausubel, F.M.,
1999. Molecular mechanisms of bacterial virulence eluci-
dated using a Pseudomonas aeruginosa – Caenorhabditis ele-
gans pathogenesis model. Cell., 96, 47–56.
[20] Tan, M., Miklos S.M., Ausubel, F.M., 1999. Killing of Cae-
norhabditis elegans by Pseudomonas aeruginosa used to model
mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci.
USA., 96, 715–720.
[21] Labrousse, A., Chauvet, S., Couillault, C., Kurz, C.L, Ew-
bank, J.J., 2000. Caenorhabditis elegans is a model host for
Salmonella typhimurium. Curr. Biol., 10, 1543–1545.
[22] Townsend, S., Hurrell, E., Forsythe, S., 2008. Virulence
studies of Enterobacter sakazakii isolates associated with a
neonatal intensive care unit outbreak. BMC Microbiol., 8,
1–9.
[23] Beale, E., Guigen, L.i., Tan M.W., Rumbaugh, K.P., 2006.
Caenorhabditis elegans senses bacterial autoinducers. Appl.
Envir. Microbiol., 72, 5135–5137.
Wyszukiwarka
Podobne podstrony:
jobm 201000013
jobm 201000298
jobm 201000191
jobm 201000321
jobm 201000018
jobm 201000214
jobm 201000067
jobm 201000037
jobm 201000074
jobm 201000280
jobm 201000385
jobm 201000198
jobm 201000458
jobm 201000147
jobm 201000520
jobm 201000327
jobm 201000342
jobm 201000420
jobm 201000364
jobm 201000317
więcej podobnych podstron